Figures & data
Table 1 MICs of the Antibiotics Tested in K. pneumoniae BKP19
Table 2 Antimicrobial Resistance Genes in K. pneumoniae BKP19
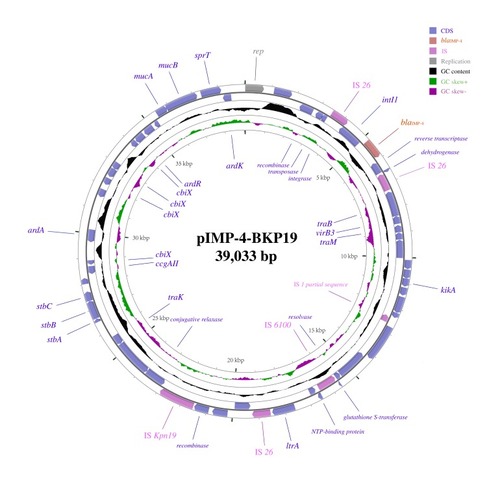
Figure 2 Sequence alignment of the pIMP-4-BKP19 plasmid from Klebsiella pneumoniae strain BKP19 with data in the NCBI GenBank database revealed several highly identical plasmids from different Gram-negative bacillus strains, including pIMP-HZ1 (K. pneumoniae strain Kp1, accession no. KU886034), p24854-IMP (K. pneumoniae strain 24854, accession no. MH909341), pIMP-GZ1058 (Escherichia coli strain CRE1058, accession no. KU051709), pIMP-HK1500 (Citrobacter freundii strain CRE1500, accession no. KT989599), pIMP-SH1506 (Enterobacter cloacae strain CRE1506, accession no. KT989598), p13SP-IMP (Klebsiella pneumoniae strain 13-sp, accession no. MH909334), P378-IMP (Pseudomonas aeruginosa strain P378, accession no. KX711879), and p128379-IMP (Enterobacter hormaechei strain 128379, accession no. MF344559).
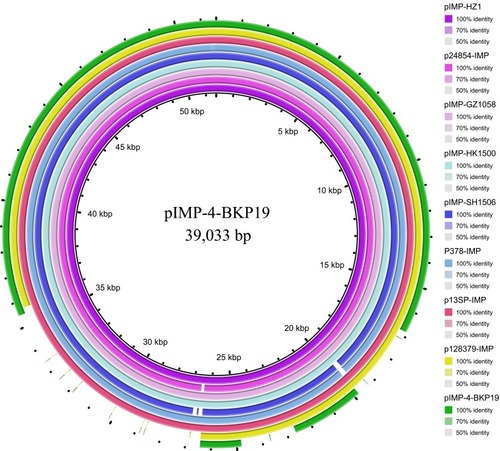
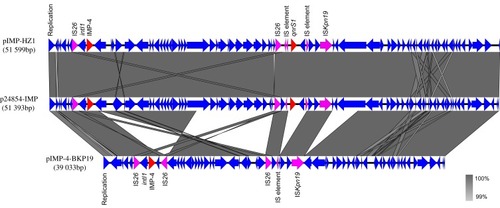
Figure 3 Comparative analysis of the homologous regions shared by three IncN-type plasmids (pIMP-4-BKP19, p24854-IMP and pIMP-HZ1). The plasmids were annotated by Rapid Annotation using Subsystem Technology (RAST) server. The figure was produced using EasyFig v. 2.2.3, and BLASTN was used to compare sequence homology with the following threshold parameters: minimum length of 100 bp accompanied by 90% identity. Antimicrobial resistance genes are indicated in red, IS elements are indicated in purple, and all other genes are indicated in blue.