Figures & data
Table 1 Characteristics of Heteroresistant Isolates Determined Using PAP, MICs of Imipenem and Levels of Biofilm Formation
Table 2 The Profile of Routine Antibacterial Resistance in P. aeruginosa
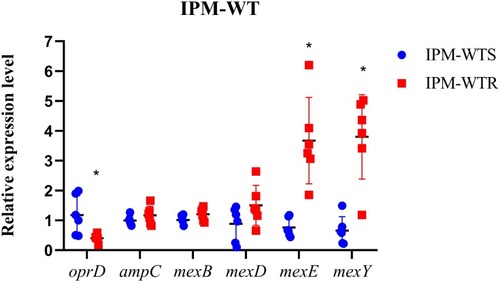
Figure 1 Levels of biofilm formation between wild-type susceptible and resistant isolates. IPM-WTS, wild-type imipenem susceptibility strains; IPM-WTR, wild-type imipenem resistance strains.
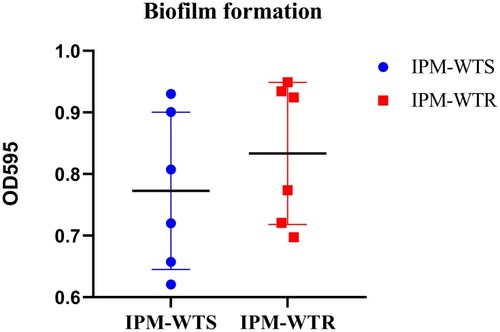
Figure 2 oprD mutations in imipenem-(hetero) resistant P. aeruginosa clinical isolates. aIn imipenem-heteroresistant populations (IPM-HRP); bwild-type imipenem resistant isolates (IPM-WTR); ctwo 3 bp deletion at nucleotide 1116–1118 and nucleotide 1147–1149 in external loop7; d1 bp deletion at nucleotide 348; e379 bp deletion at nucleotide 410–788; f1 bp deletion at nucleotide 917; g1251G > A; h15 bp deletion at nucleotide 256–270; *premature stop codon; blue circle, amino acid substitutions were detected in wild-type imipenem susceptible isolates and native imipenem susceptible populations of heteroresistance and the amino acid substitutions were predicted neutral using http://provean.jcvi.org/seq_submit.php; orange circle, oprD mutation only was detected in heteroresistance and resistance.
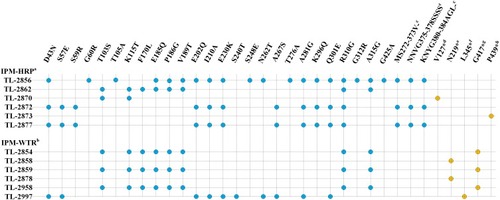
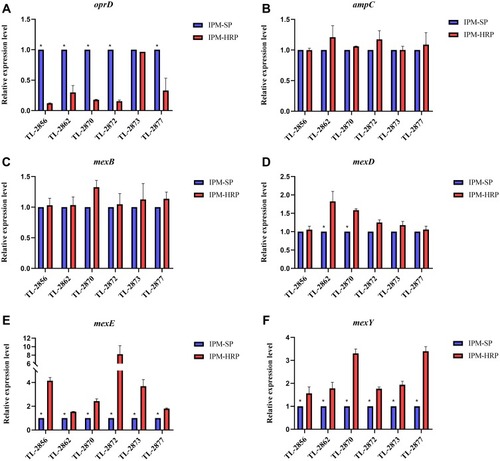
Figure 3 Expression levels of genes in heteroresistant subpopulations. (A) Expression levels of oprD. (B) Expression levels of ampC. (C) Expression levels of mexB. (D) Expression levels of mexD. (E) Expression levels of mexE. (F) Expression levels of mexY. IPM-SP, native imipenem-susceptible populations; IPM-HRP, imipenem-heteroresistant subpopulations; *P < 0.05.