Figures & data
Figure 1 Flow of sample collection and study design through the study and detection of carbapenemase-producing P. aeruginosa isolate.
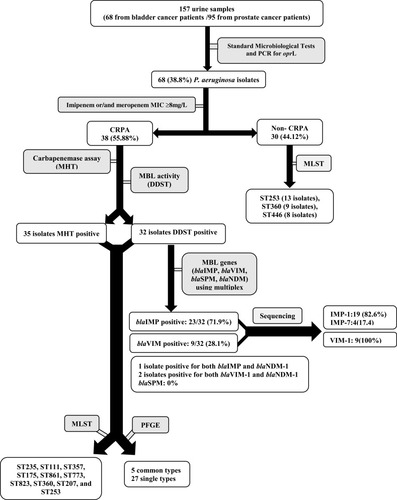
Table 1 The Susceptibility of Carbapenem-Resistant P. Aeruginosa Isolates to the Different Antimicrobial Agents
Table 2 MIC90 and MIC50 Values for Meropenem, Ceftazidime, and Cefepime Among CRPA and CSPS Isolates
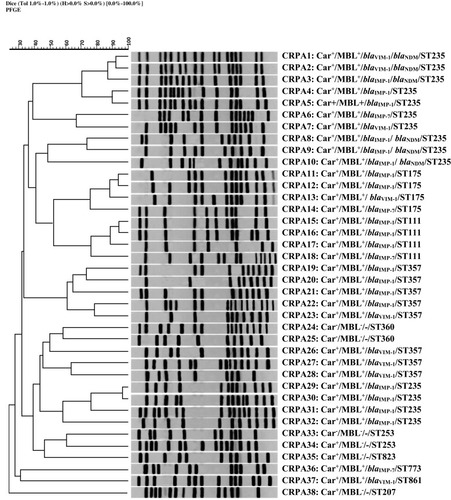
Figure 2 Genomic analysis of SpeI-digested DNA from P. aeruginosaisolates. Dendrogram based on the relationships among CRPA isolates derived from the UPGMA and Dice coefficients, using Gel ComparII software. Data about the results of MHT to identify carbapenemase activity, DDST for phenotypic detection of MBL activity, multiplex PCR assay for detection of MBL genes, and sequence type (ST) based on MLST analysis are presented, respectively.
Abbreviation: CRPA, carbapenem-resistant P. aeruginosa.