Figures & data
Table 1 Coronavirus Genera, Species and Their Binding Receptors in Hosts
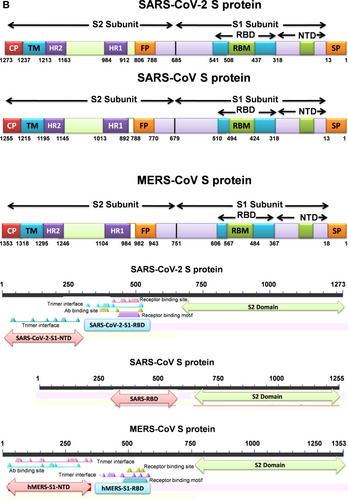
Figure 1 Graphics of the structural domains of the main coronaviruses’ spike proteins. (A) The spike glycoprotein S structure can be divided into the S1 and S2 domains, and the structural domains in the spike protein are located in the order (from C to the N terminus) as: (B) transmembrane (TM), heptad repeats (HRs) in the S2 domain, C-terminal domain (CTD), and N-terminal domain (NTD) in the S1 domain as well as the signal peptide (SP). The S1-CTD is divided into three sub-domains SD-SB, while S1-NTD contains sub-domain SA. SD-SA is accounted as receptor-binding domain (RBD).
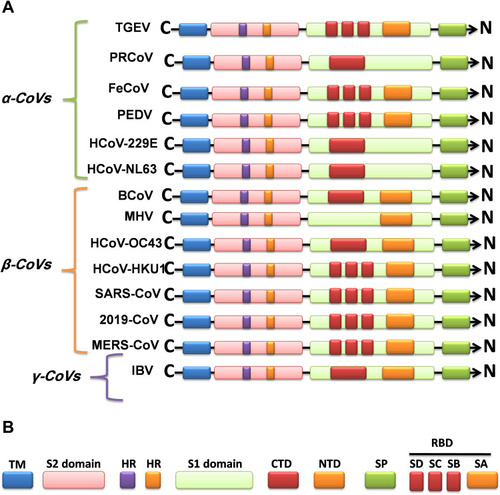
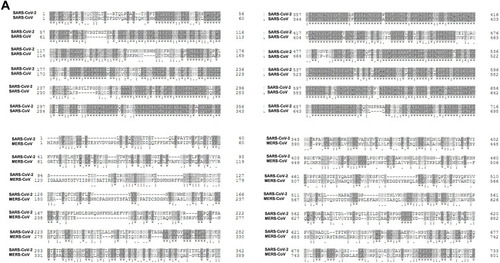
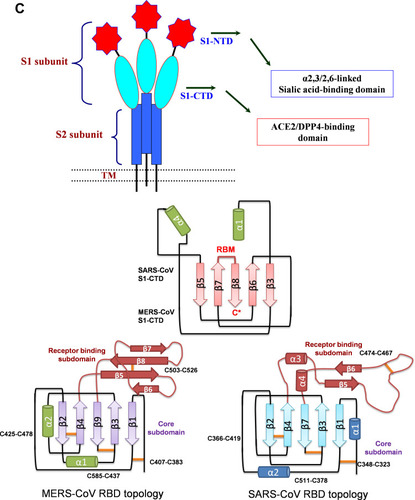
Figure 2 (A) Sequence alignment of the S protein in SARS-CoV-2, SARS-CoV and MERS-CoV, which reveals high similarities between SARS-CoV-2 and SARS-CoV S sequences shown with gray and lighted-gray. (B) Overall predicted topology of the MERS, SARS and SARS-CoV-2 spike protein, its functional domains and the related residues. The S protein mainly contains the S1 and S2 subunits and the residue numbers in each region represent their positions in the S proteins of MERDS, SARS and SARS-CoV-2, respectively. The S1/S2 cleavage sites are highlighted by simple lines. The structural domains in the spike proteins are located here in the order from the C- to the N-terminus, as: transmembrane (TM), heptad repeats (HRs) in the S2 domain, C-terminal domain (CTD), and N-terminal domain (NTD) in the S1 domain as well as the signal peptide (SP). (C) Schematic illustration of MERS-CoV and SARS RBD topologies which supposed to share some similarities. S1 contains two independent domains, an N-terminal domain (S1-NTD) and a C-terminal domain (S1-CTD). The overall structure of the SARS-CoV RBD is supposed to be similar to MERS-CoV RBD. Beta-strands are drawn as arrows and Alpha-helices are drawn as cylinders. The disulfide bonds are drawn as yellow sticks. (D) The SARS-CoV-2, SARS-CoV and MERS-CoV RBD-receptor interfaces (Contacting residues are shown as sticks at the interfaces of RBDs with receptors).
Abbreviations: SARS-CoV, severe acute respiratory syndrome coronavirus; MERS-CoV, Middle Eastern respiratory syndrome coronavirus; CP, cytoplasm domain; FP, fusion peptide; HR, heptad repeat; RBD, receptor-binding domain; RBM, receptor-binding motif; SP, signal peptide; TM, transmembrane domain/anchor; ACE2, angiotensin-converting enzyme 2; DPP4, dipeptidyl peptidase 4.
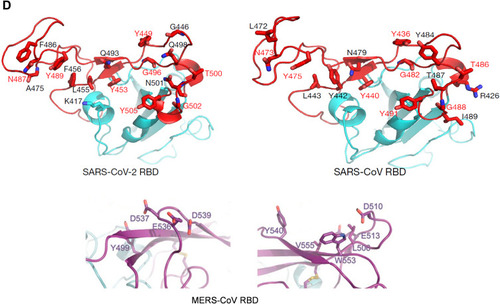
Figure 3 The hypothetical model of MERS, SARS and SARS-CoV-2 infection in the lungs of asymptomatic-to-mild (upper panel, A) and severe-to-fatal cases (lower panel, B) is illustrated here. Presented models are for two critical host determinants, eg DPP4/ACE2, cellular serine protease TMPRSS2 and sialic acid residues, differentially expressed in asymptomatic-to-mild and severe-to-fatal CoV infection. SARS-CoV-2 S engages both ACE2 and CD26 (DPP4) as the entry receptor and employs the cellular serine protease TMPRSS2 for S protein priming and efficient infectivity and spread in the host. In the entry phase, CoV S protein mediates weak interactions with abundant host surface sialates, keeping viruses concentrated on cells yet potentially diffusible across plasma membranes. S protein subsequently engages protein receptors and is proteolytically activated into membrane fusion-inducing conformations. In the Spread phase, canonical virus release is concomitant with cell-cell fusion. Cell-cell fusion involves S binding to sialic acids and does not require protein receptors, allowing infection to spread beyond the restricted distributions of protein receptors. The angiotensin-converting enzyme 2 (ACE2) in SARS-CoV and dipeptidyl peptidase 4 (DPP4) in MERS-CoV can defined as protein receptors. α2,3/2,6-SiA; α2,3/2,6-linked sialoglycans.
Table 2 A Summary of the Vaccine Development/Design, Administration Routes and Corresponding in vitro/in vivo Evaluation Results
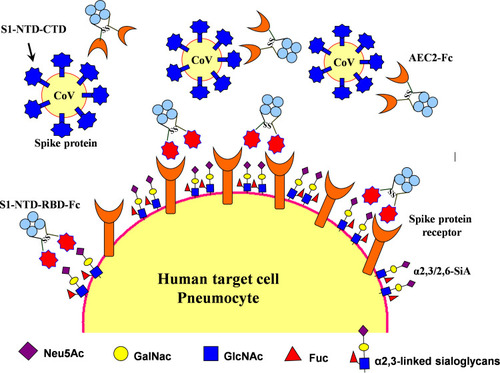
Figure 4 Therapeutic agents including S1-NTD/RBD-Fc and ACE2-Fc could be used to block 2019-nCoV from infecting cells and inducing sterilizing immunity. Target cells expressing ACE2 include lung and gastrointestinal tissues in the human body. Two proposed strategies would block this interaction and would abrogate infection. In the first, intranasal pre-infection with the S1 N-terminal domain (S1-NTD) and receptor-binding domain (RBD) of the spike protein from SARS or 2019-nCoV, attached to an human IgG1 Fc fragment (designated S1-NTD-RBD-Fc) antibody fragment (Fc), would be administered, thereby binding ACE2 and saturating available sites. Alternatively, the recombinant protein could induce sterilizing immunity. A second strategy would target the CoV virions directly by using the ACE2 extracellular domain as bait to bind to spike protein. An Fc domain fused to ACE2 would facilitate prolonged circulation of the biologic (ACE2-Fc).