Figures & data
Table 1 Demographic Characteristics, Comorbidities and Wound Burden of the Population That Was Evaluated in This Study. A Total of 28 Patients Being Treated in the TAMC Vascular Limb Salvage Clinic Were Enrolled
Table 2 Bacteria Species Recovered at Each Visit from 47 Unique Wound Sites from a Total of 28 Patients
Figure 1 A comparison of the numbers of bacterial species that were recovered from chronic non-healing wounds at the initial visit (Day 0), at a 2-week follow-up (Day 15) and at a 4-week follow-up (Day 30).
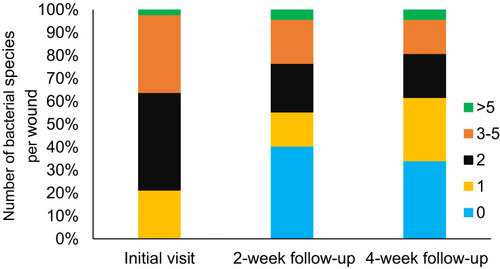
Table 3 Evaluation of the Antibacterial Resistance (AMR) Genes That Were Identified in the 123 Unique Bacterial Isolates Collected in This Study. AMR Genes Conferring Resistance to the β-Lactam Class of Antibiotic Were the Most Common, Followed by AMR Genes Conferring Resistance to Aminoglycosides and the Macrolides, Respectively
Table 4 A Summary of Antibacterial Resistance (AMR) Detections by Bacterial Species and Potential Impact on Antibiotic EffectivenesS. The Members of the Enterobacteriaceae (E. Coli), The Non-Fermenters (P. Aeruginosa), and the S. Aureus Were Found to Harbor the Greatest Numbers of AMR Genes
Table 5 Phenotype Conversion of S. Aureus from Sensitive (S) to Resistant (R) for Clindamycin and Erythromycin Over a Period of 30 Days
Table 6 Single Nucleotide Polymorphisms (SNPs) Detected in Three Isolates of S. Aureus Recovered from Wound Sites on a Single Patient on Days 0, 15 and 30. All SNPs Were Localized to Genes That Have Been Predicted to Code for Proteins Involved in the Metabolic Process of the Pathogen and No SNPs Were Detected in Any of the Genes Known to Be Involved in the Development of Antibiotic Resistance
