Figures & data
Table 1 Clinical Characteristics of Patients Infected with CRKP and hvKP
Table 2 Demographics of Patients Infected with CRKP and hvKP
Table 3 Antimicrobial Susceptibility Testing Results of 12 K. pneumoniae Isolates (Mg/L)
Table 4 Distribution of Resistance Mechanism and Virulence Genes Harbored by CRKP and hvKP Strains
Figure 2 Distribution of MICs with NDM-1, NDM-5 and KPC-2/CTX-M-14.
Notes: MIC distribution of ATM, quinolones, and TGC in blaNDM-1 (n = 3), blaNDM-5 (n =1), and blaKPC-2/CTX-M-14 (n = 1) CRKP strains.
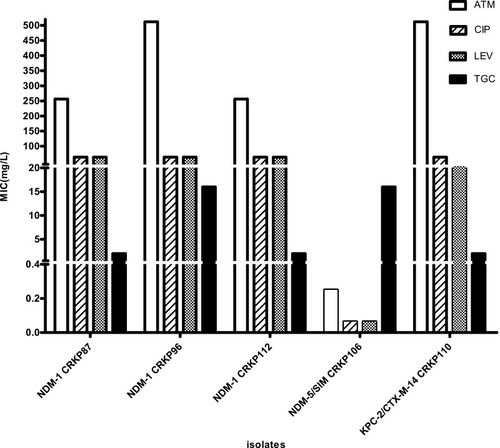
Figure 3 Molecular phylogenetic analysis of 12 isolates.
Notes: The evolutionary history by using maximum likelihood method based on the Kimura-2-parameter model; evolutionary analyses were conducted in MEGA-X.
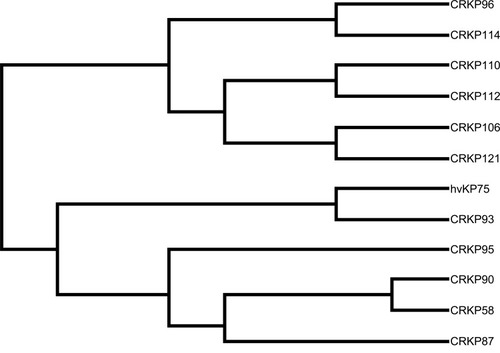
Table 5 The Distribution of Resistance Genes
Table 6 The Distribution of Virulence Genes
Table 7 The FICs of CRKP Isolates