Figures & data
Figure 1 Phylogenetic analysis of 135 isolates in this study. The branches of different CT type isolates are shown in colours. The corresponding isolates of each CT type are CT1291 (P1291-1 to P1291-20), CT1313 (P1313-1 to P1313-33, AH1313-1 to AH1313-2), CT1689 (AH1689-1 to AH1689-8), CT1772 (AH1772-1 to AH1772-4), CT1814 (AH1814-1 to AH1814-13), CT1819 (AH1819), CT2405 (P2405-1 to P2405-9), CT2410 (P2410-1 to P2410-11), CT2418 (P2418-1 to P2418-4), CT2444 (P2444), CT2445 (P2445-1 to P2445-6), CT3175 (AH3175-1 to AH3175-2), CT3176 (AH3176-1 to AH3176-14), CT3178 (AH3178-1 to AH3178-4), CT3179 (AH3179-1 to AH3179-2), and CT3195 (AH3195). rmpA, rmpA2, magA iroB, peg-344 and iucA were identified for the differentiation of hypervirulent K. pneumoniae from classical K. pneumoniae in a previous study.Citation18
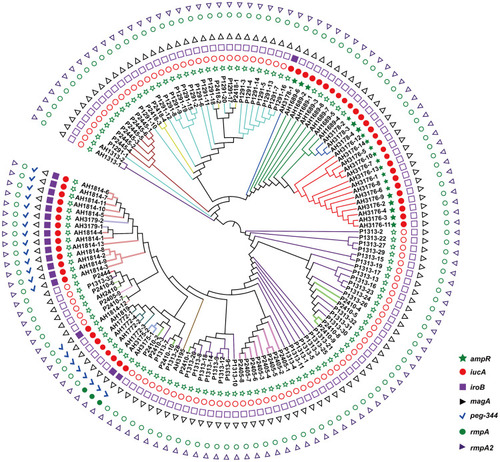
Table 1 Demographics, Comorbidities, Invasive Procedures and Mortality of Patients with Carbapenem-Resistant Klebsiella pneumoniae Infections
Figure 2 Pan-GWAS analysis between the CT3176 and non-CT3176 groups. The pangenome and associations were calculated with Roary and Scoary. A plot map was generated with ggplot2.
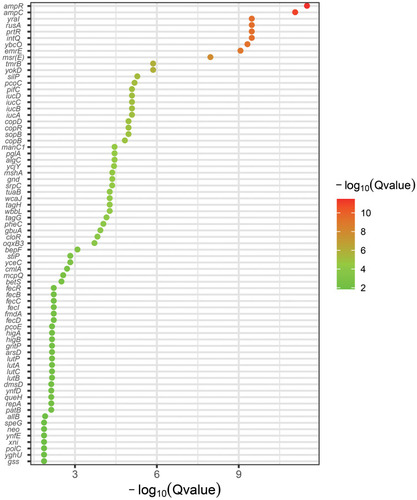
Figure 3 Proteome analysis and capsule staining. (A) Volcano plot showing the comparison of quantitative protein expression between ampR+ (AH1689-2, AH3176-1 and AH3178-1) and ampR− (AH1689-3, AH1689-4, and AH1689-5) Klebsiella pneumoniae isolates. (B, C) Differentially expressed proteins with a fold change over 1.2 are marked in colour.
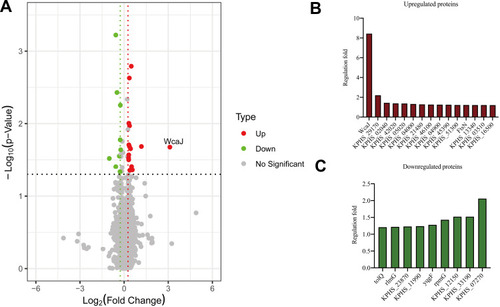
Figure 4 Virulence potential of non-hypermucoviscous Klebsiella pneumoniae isolates in a mouse pulmonary infection model. (A) The effect of 1 × 10Citation7 colony-forming units (CFU) of AH3176-1, AH1689-2, AH1689-3 and AH1689-3 ampRKL47 complement mutant on survival was assessed. (B) At 24 hours post-infection (hpi), the lungs of infected mice were harvested, and the bacterial burden was determined by CFU enumeration (n = 10; *P < 0.05, **P < 0.01, one-way ANOVA). Data are representative of 3 independent experiments. (C) Uronic acid production of different strains. An unpaired two-sided Student’s t-test was performed. Each data point was repeated three times (n = 3). Data are presented as the mean ± s.e.m. *P < 0.05, ns = no significance. The lungs of mice without infection (D) or infected with ATCC700721 (E) and AH3176-1 (F) were collected and stained with haematoxylin and eosin. All images are at ×40 magnification. Scale bars: 250 μM.
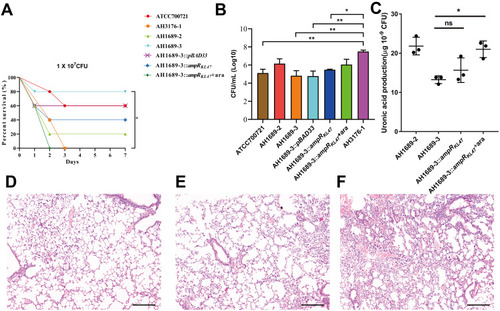
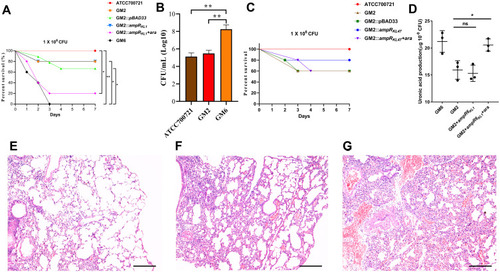
Figure 5 Virulence potential of hypermucoviscous Klebsiella pneumoniae isolates in a mouse pulmonary infection model. (A) The effect of 1 × 106 colony-forming units (CFU) of GM2, GM6 and GM2 ampRKL1 complement mutant on survival was assessed (n = 10; *P < 0.05, **P < 0.01, log-rank Mantel-Cox test),). (B) At 24 hours post-infection (hpi), the lungs of infected mice were harvested, and the bacterial burden was determined by CFU enumeration (n = 10; **P < 0.01, one-way ANOVA). Data are representative of 3 independent experiments. (C) The effect of 1 × 106 CFU of GM2 and GM2 ampRKL47 complement mutant on survival was assessed. (D) Uronic acid production of different strains. An unpaired two-sided Student’s t-test was performed. Each data point was repeated three times (n = 3). Data are presented as the mean ± s.e.m. *P < 0.05, ns = no significance. The lungs of mice without infection (E) or infected with GM2 (F) and GM6 (G) were collected and stained with haematoxylin and eosin. All images are at ×40 magnification. Scale bars: 250 μM.