Figures & data
Table 1 Primer List for Next-Generation Sequencing Studies
Table 2 Demographic Data of the Treatment-Naïve HIV-1-Infected Patients (n=224)
Table 3 INSTI Drug Resistance and Resistance-Associated Mutations by Sanger Sequencing Among Treatment-Naïve HIV-1-Infected Patients (n=224)
Table 4 Demographic Data of the HIV-1-Infected Patients Who Underwent Both Sanger Sequencing and NGS for INSTI Resistance (n=38)
Table 5 Overall Frequency (%) of INSTI Resistance-Associated Mutations Among HIV-1-Infected Patients by NGS (n=38)a
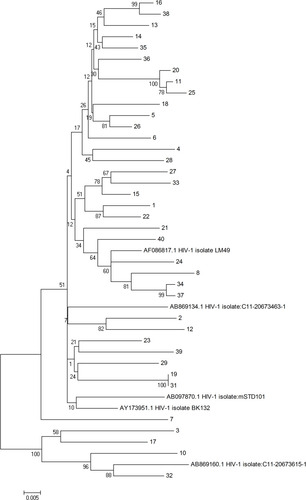
Figure 1 Percentage of HIV drug resistance and resistance-associated mutations to NRTIs, NNRTIs, PIs and INSTIs among 224 HIV-1-infected patients with GRT by sanger sequencing. The figure shows that a 8.9% had drug resistance to any of the four classes of anti-retroviral drugs, including 4% who had resistance to NRTIs, 5.8% to NNRTIs, 0.4% to PIs and 0.9% to INSTIs.
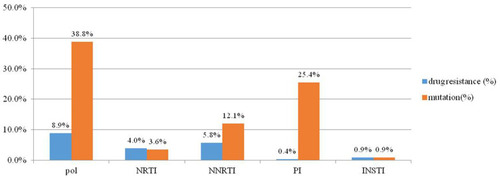
Figure 2 The prevalence of drug resistance to NRTIs, NNRTIs, PIs and INSTIs among 224 HIV-1-infected treatment-naïve patients in southern Taiwan. The figure shows that 2.7% had resistance to lamivudine, emtricitabine, abacavir and stavudine, 1.8% had resistance to tenofovir, 5.8% had resistance to nevirapine, 4.9% had resistance to efavirenz, and 0.9% had resistance to raltegravir and elvitegravir.
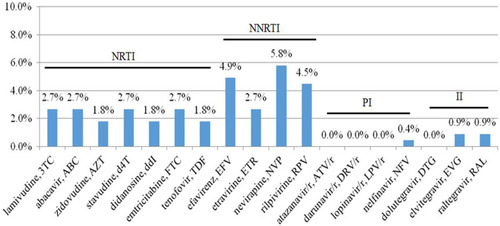
Figure 3 The prevalence of drug resistance-associated mutations to NRTIs, NNRTIs, PIs and INSTIs among 224 HIV-1-infected patients with transmitted drug resistance. The figure shows that the most common drug resistance-associated mutations were M184V (1.3%) and K65R (1.3%) for NRTIs, V179D (4.5%), V106I (2.7%) and K103N (1.3%) for NNRTIs, and L10I (13.4%) and A71T (5.8%) for PIs. Overall, 0.4% of the patients had drug resistance-associated mutations to G163K and E138A.
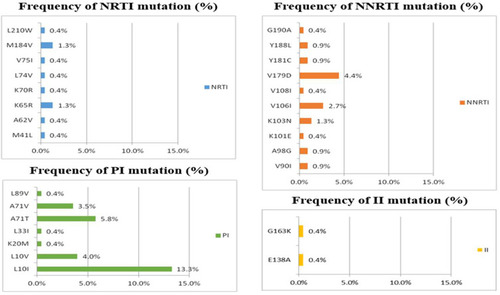
Figure 4 Phylogeny from 38 patients enrolled from the 2017 VCT program who received NGS and sanger sequencing were inferred from the integrase gene sequences. (Subtype B reference strain: AB869134, AF86817, AB097870, AY173951. Subtype 01AE reference strain: AB869160). One cluster was detected (sample 19 and 31).