Figures & data
Table 1 Primers Designed for Gene-PCR of the gFlHb of Giardia lamblia Sub-Assemblage AII and Assemblage B All Primers Were Ordered from Eurofins Genomics (Ebersberg, Germany)
Table 2 Estimates of gFlHb Copy Numbers in PacBio and Illumina Sequencing Data, Number of Clones, and Identified Alleles and SNVs in Cloned Sequences
Figure 1 Phylogenetic tree of all the alleles of gFlHb found in the sub-assemblage AII isolates and gFlHb copies found in the PacBio sequencing data. A is abbreviated for allele, and the number of clones representing each allele is listed in the parenthesis.
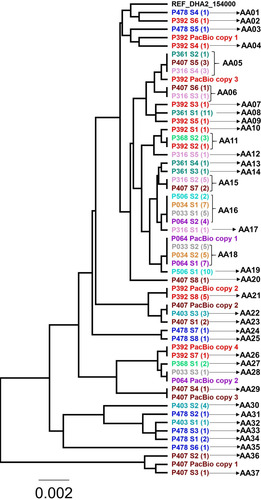
Figure 2 Phylogenetic tree of all the alleles found in the assemblage B isolates and gFlHb copies found by PacBio sequencing. A is an abbreviation of allele, and the number of clones representing each allele is listed in the parenthesis.

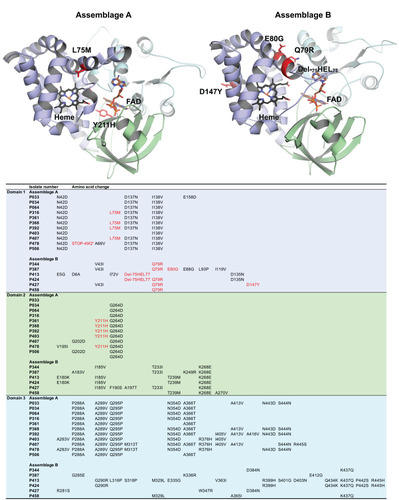
Figure 3 Cartoon representation of homology models created for gFlHb from assemblage A and B. Heme binding globing domain is presented in purple, FAD-binding domain in green and C-terminal FAD- binding domain in cyan. Heme and FAD were fitted to the homology model and are presented by stick representation. Detected nsSNV-induced mutations to the amino acid sequences for individual isolates are presented in the table. Mutations possibly affecting directly to protein function by disturbing the heme or FAD binding are indicated with red in both cartoon models and in the table. The mutation marked as STOP-49Q introduces a stop codon into the sequence interrupting the protein synthesis.