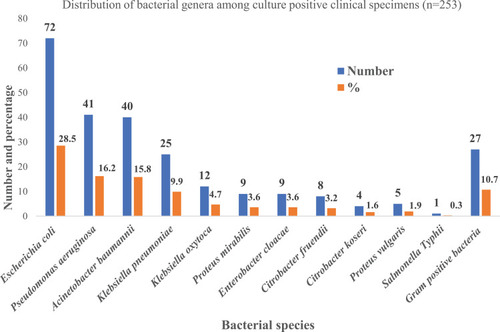
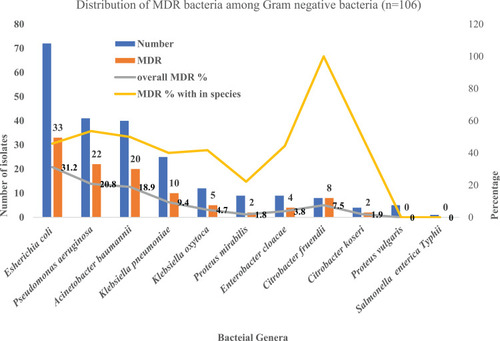
Figures & data
Table 1 Demographic and Clinical Character of Patients Attending at Annapurna Neurological Institute and Allied Sciences
Table 2 Antibiotic Susceptibility Pattern of Gram-Negative Bacterial Isolates (n=226)
Figure 3 Agarose gel electrophoresis (1.5%) used for separation of PCR products. Lane 2, positive control; Lane 3, 5, and 7 are CITM positive; Lane 4 and 6, CITM negative; and Lane 8. negative control.
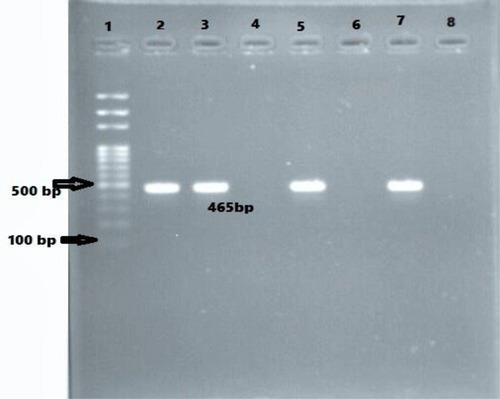
Figure 4 Agarose gel electrophoresis (1.5%) used for separation of PCR products. Lane 2, positive control; Lane 3, 4, 6 and 7 are Dham positive; Lane 5, Dham negative; and Lane 8, negative control.
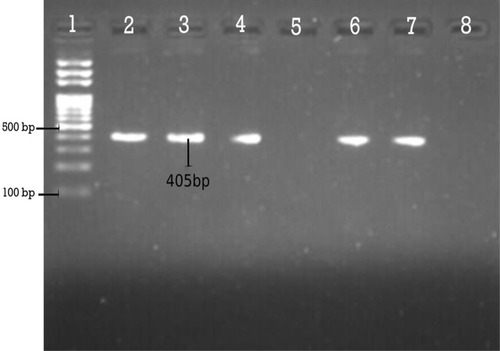
Table 3 Distribution of AmpC β-Lactamase, CITM and DHAM Genes Among Gram-Negative Bacterial Isolates and Their Relation to Gender, Age and Clinical Specimens