Figures & data
Figure 1 Minimum spanning tree generated by cgMLST coloured by ST (Pasteur) based on 3487 targets. Numbers between the nodes indicate the number of alleles that are different.
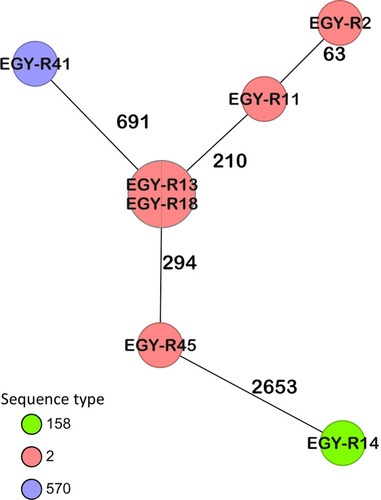
Table 1 Resistomes in Seven Colistin-Resistant A. Baumannii Isolates by WGS
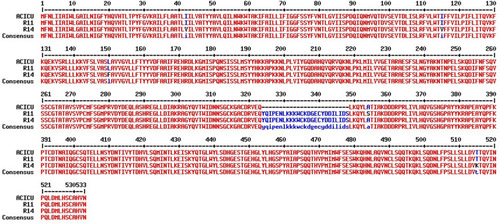
Table 2 Amino Acids Substitution in PmrCAB System in Seven Sequenced Isolates in Comparison to ACICU Strain
Data availability
Repositories:
Raw reads generated were submitted to the Sequencing Read Archive (https://www.ncbi.nlm.nih.gov/sra/) of the National Center for Biotechnology Information (NCBI) under the SRA accession number SRP131448 and BioProject PRJNA431710.