Figures & data
Table 1 Antibiotic susceptibility profiles of A. hydrophila SS332 and transformants
Table 2 General features of Aeromonas hydrophila SS332 genome
Figure 1 Circular map of complete genome sequences of pSS332-218k and comparison with pMCR5–045096. Circles 1-5 (from inside to outside) represent the information as follows: (1) GC content, (2) GC skew, (3) negative-strand CDSs, (4) positive-strand CDSs and (5) pMCR5-045096. Functional features of pSS332-218k are highlighted in different colors (mobile genetic elements are in red; antimicrobial resistance genes are in yellow; the other known genes are in indigo blue; and hypothetical protein genes are in gray).
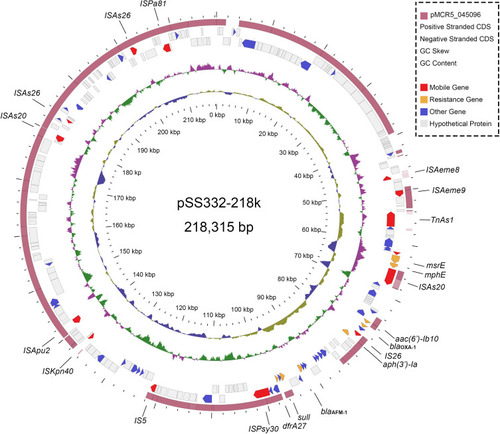
Figure 2 Phylogenetic analysis and sequence alignment of AFM-1. (A) Phylogenetic analysis of the protein sequence using the maximum likelihood method. (B) Sequence alignment analysis of AFM-1 and representative MBLs of subclass B1. Residue numbers are positioned above the sequences according to the class B β-lactamase standard numbering scheme; black spots indicate the conserved zinc-binding residues at H117, H119, D121, H186, C205 and H247. Green spots represent the essential residues at D88 and D189 for the stable expression of NDM β-lactamase.
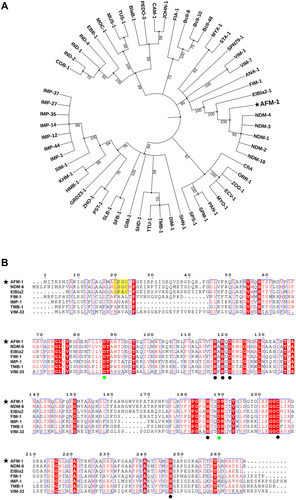
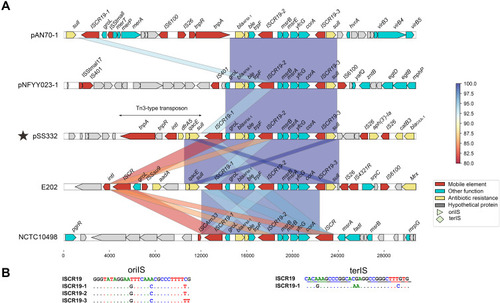
Figure 3 BLAST homology search results. (A) The homology of the whole nucleotide sequence of blaAFM-1 was generated against the nr/nt database using BLASTN. The GenBank accession numbers for the sequences are as follows: Alcaligenes faecalis AN70 plasmid pAN70-1, MK757441.1; Comamonas testosteroni pNFYY023-1, MT11984.1; Bordetella trematum E202, CP049957.1; Stenotrophomonas maltophilia NCTC10498, CP049956.1. (B) Three novel ISCR19-like elements, ISCR19-1, ISCR19-2 and ∆ISCR19-3.