Figures & data
Table 1 Specific PCR Primer Pairs Used in This Study.
Figure 1 PFGE patterns from 19 ESBL-positive E. coli isolates obtained from an NICU outbreak of infection from Ha’il, Saudi Arabia in 2014. Cluster analysis was performed using the method of DICE with UPGMA with band tolerances set to 1.0%. MIC values (µg/mL): amikacin (AK) S = ≤16; gentamicin (GM) S = ≤4; tobramycin (TOB) S ≤4, I = 8; ampicillin (AMP) S ≤8, R ≥32; cefoxitin (CFT) R ≥32; ceftazidime (CAZ) S ≤4, R ≥16; ciprofloxacin (CIP) S ≤0.25; levofloxacin (LEVO) S ≤0.5, imipenem (IMI) S ≤1. Abbreviations: MLST, 131 - Multilocus sequence type 131. TEM, N - no TEM gene detected; 1 - TEM 1 detected. SHV, N - no SHV gene detected; 2 - SHV 2 detected; 12 - SHV 12 detected. CTX-M, 15 - CTX-M-15 detected.
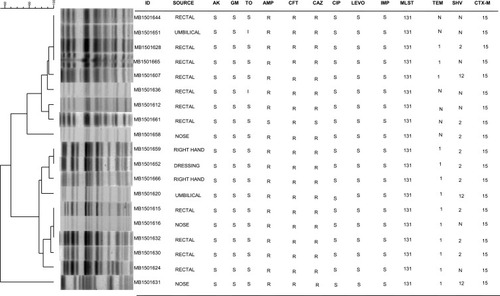
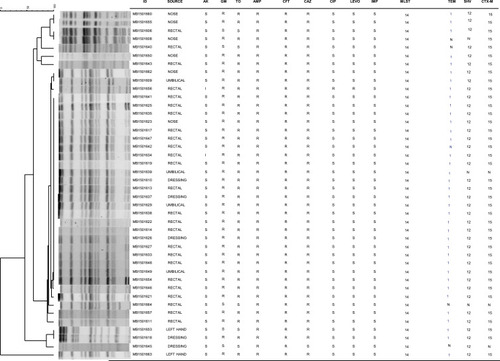
Figure 2 PFGE patterns from 42 ESBL-positive K. pneumoniae isolates obtained from an NICU outbreak of infection from Ha’il, Saudi Arabia in 2014. Cluster analysis was performed using the method of DICE with UPGMA with band tolerances set to 1.0%. MIC values (µg/mL): amikacin (AK) S ≤16, I = 32; gentamicin (GM) S ≤4, R ≥16; tobramycin (TOB) S ≤4, I = 8, R ≥16; ampicillin (AMP) R ≥32; cefoxitin (CFT) R ≥32; ceftazidime (CAZ) S ≤4, R ≥16; ciprofloxacin (CIP) S ≤0.25, R ≥1; levofloxacin (LEVO) S ≤0.5, R ≥2; imipenem (IMI) S ≤1. Abbreviations: MLST, 14 - Multilocus sequence type 14. TEM, N - no TEM gene detected; 1 - TEM 1 detected. SHV, N - no SHV gene detected; 12 - SHV 12 detected. CTX-M, N - no CTX-M gene detected; 15 - CTX-M-15 detected.