Figures & data
Figure 1 Outline of the Ohashi Medical Center, Toho University. Prior to relocation to the new building (BN), the hospital consisted of four buildings: West building (BW), Administration building (BA), Central building (BC), and East building (BE). Each building had respective sewage tanks: BW: STW, BA: STA, BC: STC, BE: STE, BN: STN.
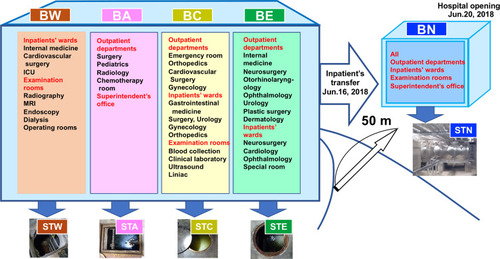
Table 1 Concentrations of Chemical Compounds in the Hospital Sewage Tank
Figure 2 PCoA plot based on NGS read counts detected by metagenome DNA-Seq. PCoA was performed according to Bray Curtis distance (the average linkage). The genera, Acinetobacter, Citrobacter, and Comamonas were frequently detected in STW and STN samples. Most severely ill inpatients were treated in the BW and BN; thus, their excretion may have a major impact on the bacterial content of the sewage tanks.
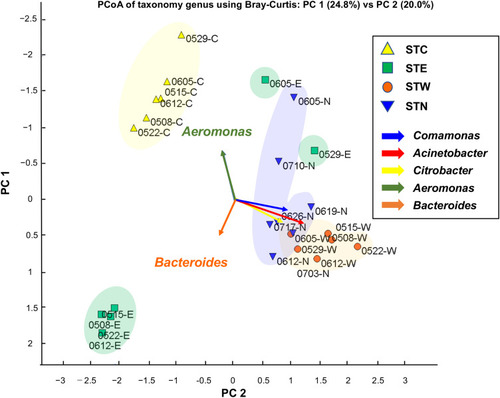
Figure 3 Core genome phylogeny using single-nucleotide variations (SNVs) of ESBL-producing E. coli isolates. Core genome phylogeny was constructed using ESBL-producing E. coli isolates; 20 clinical isolates (THO-number, orange highlighted), one sewage isolate from STW0522 (brown highlighted), and 20 sewage isolates from STN0717 (blue highlighted). The complete genome sequence of STN0717-11 was used as a genome reference and 39.48% of the genome sequence was used as core genome regions among all tested strains. Few clinical isolates were obtained from same patient (§, patient No.8; ¶, patient No.9; †, patient No.7 in Supplement Data Set S2). Heatmap of pairwise differences of core genome SNVs are shown using a colour gradient with pink and red. The lower half part indicates core genome SNVs among all strains, and the upper half part shows core genome SNVs between indicated two strains. THO-008 and -019 from the same patient showed no SNVs with sewage isolates (14 isolates; STN0717-1 to −11, 14, 15, and 19). There were no identical clones between different patients.

Table 2 ESBL-Producer in Hospital Sewage and Clinical Isolate
