Figures & data
Table 1 Socio-Demographic and Clinical Characteristics of the Study Subjects Among First Line ART Experienced Patients at South Omo, Ethiopia (n=253)
Figure 1 Phylogenetic tree of 41 HIV samples collected from South Omo Zone, Ethiopia. cDNA was prepared from HIV-1 genomic RNA, and a region spanning codons 90–234 of HIV reverse transcriptase was PCR amplified and sequenced on the Illumina MiSeq platform. A neighbor-joining treeCitation30,Citation32 constructed from the consensus sequences is depicted as a circular cladogram. Clinical samples are coded by a prefix OMO, indicating the name of the study site followed by two digit numbers. HIV-1 subtype consensus sequences (n=25) spanning RT codons 90 to 234 were included and represent subtypes A1, A2, B, C, D, F1, F2, G, and H, as well as recombinant viruses AE, AG, AB, BC, CD, BF, BG (retrieved from http://www.hiv.lanl.gov) and reference amplicon. HIV-1 RT sequences were primarily subtypes C. There were a total of 476 positions in the final dataset. Evolutionary analyses were conducted in MEGA6Citation27.
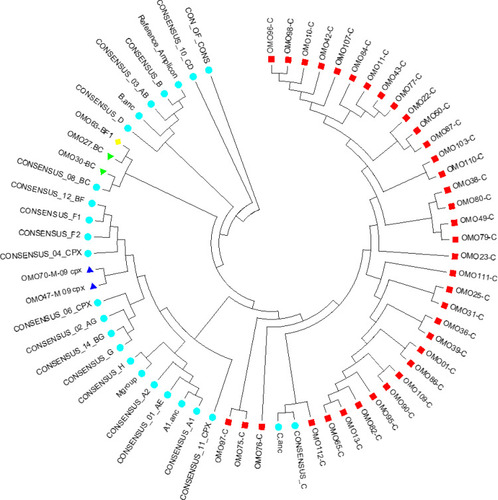
Figure 2 ADR mutation frequency at 5%and 20% detection sensitivity thresholds, as determined by MiSeq sequencing.
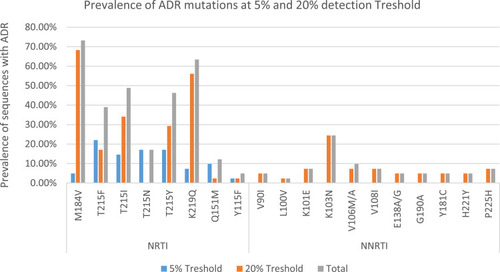
Figure 3 Predicted ARV drug responses of 35 patients with ADR mutations. Drug responses were based on the Stanford HIVdb. The y-axis indicates the number of sequences with ADR while the x-axis indicates the different ARV druelsg classes that were affected by the ADR mutations.
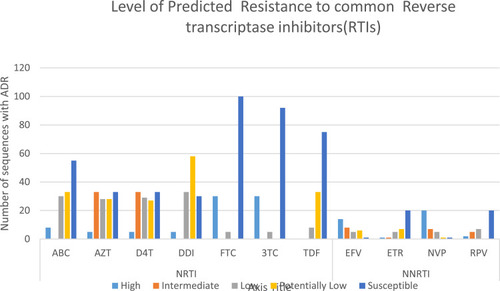
Table 2 HIV-1 Drug Resistance Conferring Mutations in the RT Gene Among First Line ART Experienced Adults with ADR Mutations, South Omo, Ethiopia, 2017 (N=41)
Table 3 Logistic Regression Analysis Describing Associations of HIV-1 Drug Resistance Mutations Among First Line ART Experienced Patients in South Omo, Ethiopia (n=253)
