Figures & data
Table 1 The Primer Sequences for Diagnosis of Bacterial Isolates BM Patients
Table 2 Demographics of Bacterial Meningitis Infected Patients
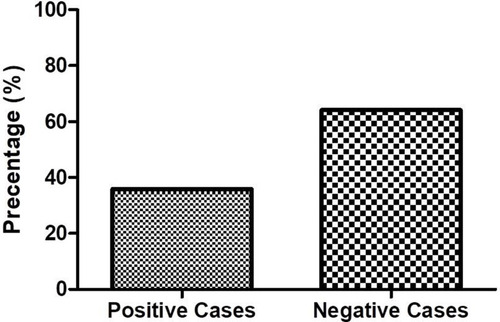
Table 3 Comparative Analysis of Bacterial Culture and Gram Staining Among Bacterial Meningitis Cases
Table 4 Incidence Rate of Isolated Bacterial Meningitis Pathogens Among Various Age Groups
Table 5 Prognostic Factors of Bacterial Meningitis Infected Patients
Figure 3 16S rRNA gene PCR-based identification bacteria isolated from CSF of bacterial meningitis patients. (A) E. coli isolates with 204bp amplified fragments; (B) S. aureus solates 257bp amplified fragments; (C) K. pneumoniae isolates 525bp amplified fragments; (D) S. pneumoniae isolates 598bp amplified fragments; (E) H. influenzae isolates 528bp amplified fragments; (F) N. meningitidis isolates 568bp amplified fragments. M= molecular weight marker (1kb).
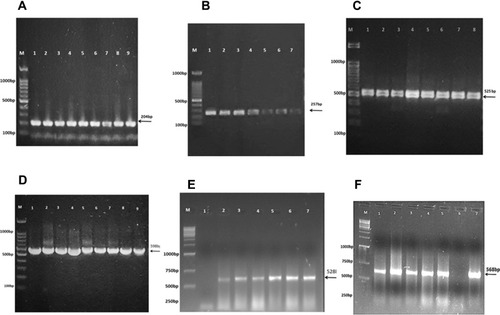
Figure 4 Phylogenetic relationship of bacterial isolates from bacterial meningitis patients with their reference strains. S. pneumoniae (MW405781, MW405782, MW405783, MW405784, MW405785, MT75569.1); E. coli (MT75572 MT916768, MT916769, MT916770); K. pneumoniae (MW208813, MW208814, MW208815, MW208816, MW208817, MT102939); S. aureus (MW148201.1, MW148202.1, MW148203.1, MW148204.1, MT75573.1); H. influenzae (MZ348556, MZ348557, MZ348558, MZ348559, MZ348560); N. meningitidis (MZ318247, MZ318248, MZ318249, MZ318250, MZ318251) isolated from BM patients of Balochistan were compared using 16S rRNA gene sequence with other reference strains using neighbor-joining phylogenetic tree. The bootstrap values were based on 1000 replicates in MEGA X software.
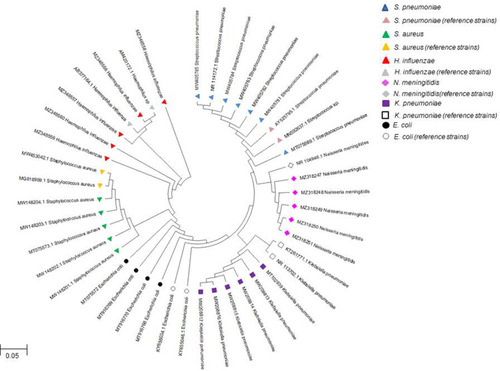
Table 6 The Antimicrobial Susceptibility Test Against Bacteria Isolated Bacterial Meningitis Patients