Figures & data
Figure 1 Characteristics of IPM heteroresistance among the Kp19, Kp25 and Kp34 multidrug-resistant K. pneumoniae isolates. (A) Satellite colonies in the IPM MIC gradient strips (black arrows) for three K. pneumoniae isolates; (B) population analysis profiles of the three multidrug-resistant K. pneumoniae and control strain; (C) workflow for subculture of IPM-susceptible and -resistant subpopulations. Cultures of Kp19, Kp25 and Kp34 were grown for 18 h in medium containing 8 μg/mL imipenem or drug-free medium, respectively; (D) PFGE results of the imipenem-resistant (RS) and -susceptible subpopulations (SS) of heteroresistant strains. IPM, imipenem; ATCC13883, K. pneumoniae ATCC13883.
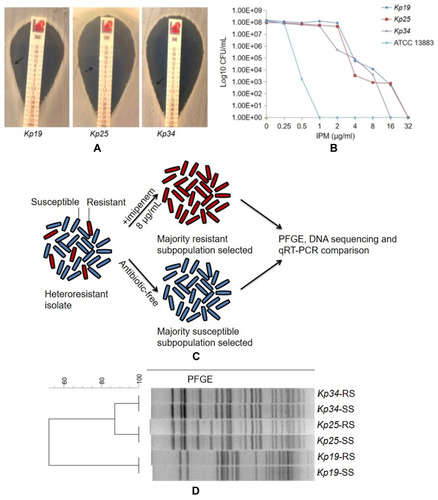
Table 1 Phenotypic and Molecular Characterisation of 37 Multidrug-Resistant K. pneumoniae Clinical Isolates
Table 2 Analysis of the Presence of IPM Heteroresistance in Three MDR K. pneumoniae Isolates and Sequencing Profiles of ompK35 and ompK36 Genes
Figure 2 Heteroresistant K. pneumoniae isolates lead to IPM treatment failure in vivo. Mice were infected with the IPM-susceptible K. pneumoniae ATCC13883 or the IPM-heteroresistant isolates, treated with imipenem/cilastatin sodium or PBS at 12 h and 18 h after infection, and then euthanized at 24 h and the peritoneal lavage fluid was collected. (A) Numbers of CFU were quantified at 24 h in the peritoneal lavage fluid; (B) increase in the frequency of the IPM-resistant subpopulation in the peritoneal lavage fluid; (C) survival of mice infected with the K. pneumoniae ATCC13883 and then treated with imipenem or PBS; (D) survival of mice infected with the IPM-heteroresistant Kp19 and then treated with imipenem or PBS; (E) survival of mice infected with the IPM-heteroresistant Kp25 and then treated with imipenem or PBS; (F) survival of mice infected with the IPM-heteroresistant Kp34 and then treated with IPM or PBS. Surviving mice were monitored until day 3 (n = 5), Error bars represent SEM (Mann–Whitney test). VF, virulence factor; ATCC13883, K. pneumoniae ATCC13883; *P values are significant (P <0.05).
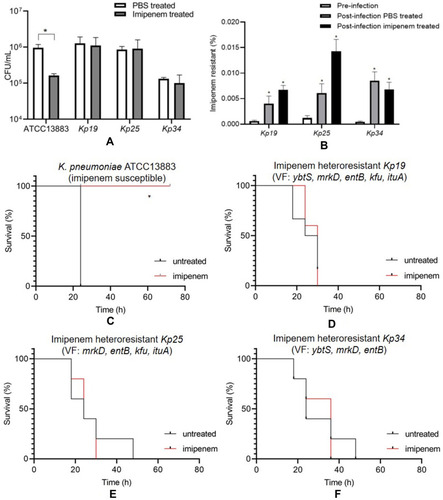
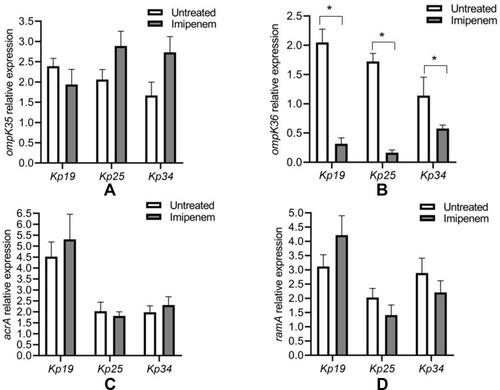
Figure 3 Relative fold gene expressions in the IPM-resistant and -susceptible subpopulations (untreated) of heteroresistant K. pneumoniae isolates. (A) Relative expression of ompK35 gene; (B) relative expression of ompK36 gene; (C) relative expression of acrA gene; (D) relative expression of ramA gene. *P values are significant (P <0.05).