Figures & data
Table 1 Target Genes, Sequences of PCR Oligonucleotide Primers and Expected PCR Product Size
Table 2 Distribution of Antimicrobial Resistance Among Gut Commensal E. coli Isolates
Figure 1 Representative agarose gel (0.7%) electrophoresis of the different patterns of extracted plasmids from gut commensal E. coli isolates. The patterns comprised of one band (lanes 2, 3, 6, 8, 10, 12), two bands (lanes 1, 5, 7, 9, 11) and three bands (lane 4) of different sizes. First most left lane, 1 kb DNA molecular weight marker.
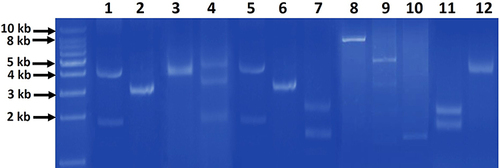
Table 3 Distribution of AMR Genes with Corresponding AMR Phenotypes Detected in E. coli Isolates
Figure 2 Prevalence of AMR-phenotypes and associated genes in commensal E. coli isolates. (A) AMR phenotypes among E. coli isolates; (B) AMR-associated genes harbored by E. coli resistant phenotypes.
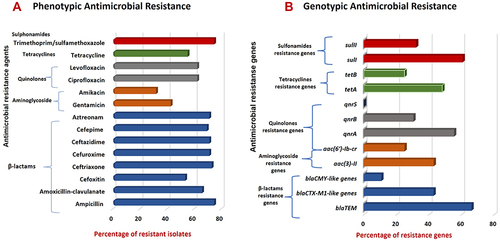
Table 4 Genotypic Profiles of AMR Genes Among E. coli Isolates
