Figures & data
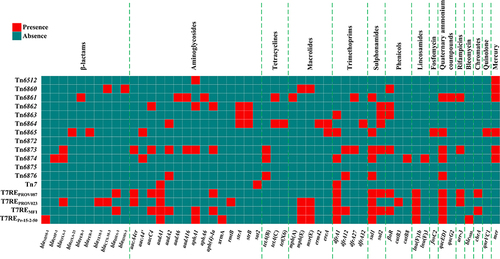
Table 1 Major Features of the 17 AGEs Characterized in This Work
Figure 2 Massive gene acquisition and loss across whole-genomes of seven related ICEs Tn6512, Tn6860, Tn6861, Tn6862, Tn6863, Tn6864, and Tn6865.

Figure 3 Organization of Tn6896 from Tn6864, and comparison to related regions.
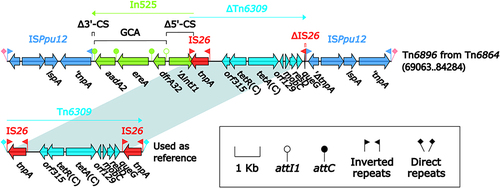
Figure 4 Comparison of MDR regions from Tn6860, Tn6861, Tn6862, Tn6863, Tn6864, and Tn6865.
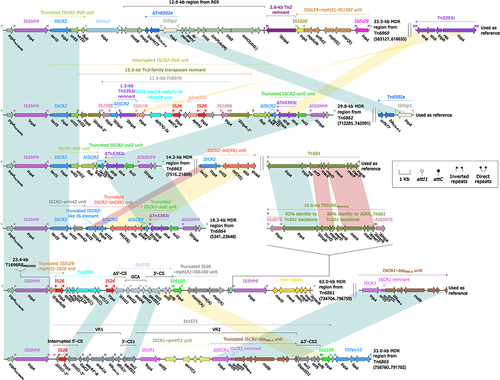
Figure 5 Comparison of three related IMEs Tn6872, Tn6873, and Tn6874.
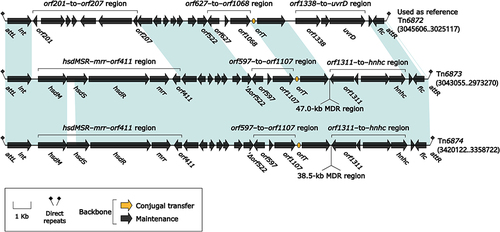
Figure 6 Organization of MDR regions from Tn6873 and Tn6874, and comparison to related regions.
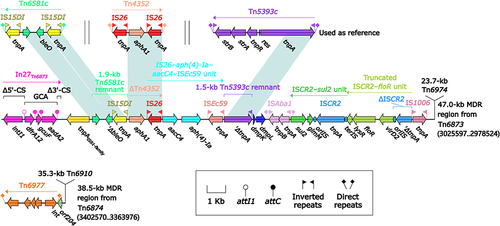
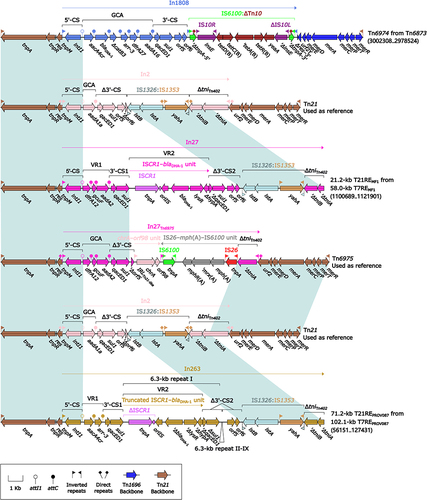
Figure 7 Comparison of two related IMEs Tn6875 and Tn6876.
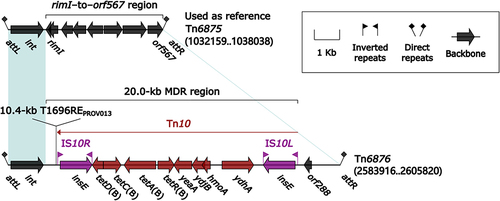
Figure 8 Comparison of Tn7 and its four derivatives.
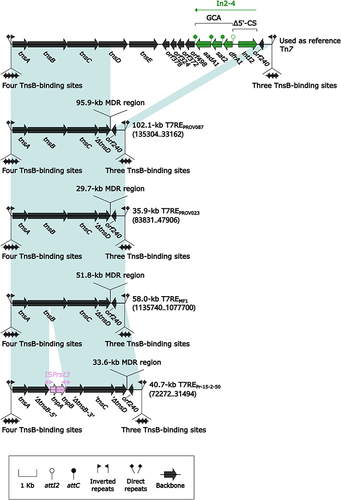
Figure 9 Comparison of MDR regions from four Tn7 derivatives.
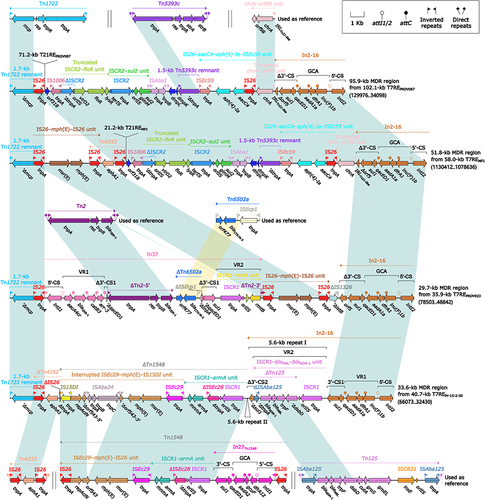
Figure 10 Comparison of Tn1696 and its three derivatives.
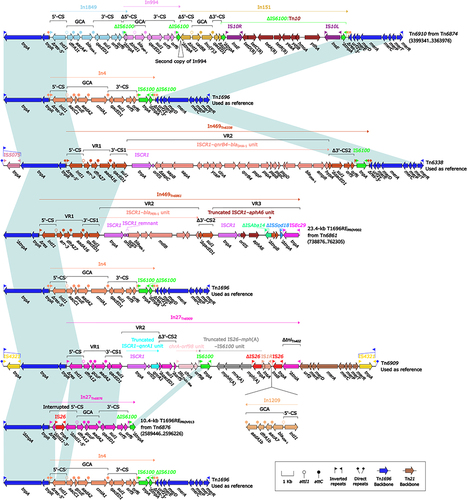
Figure 11 Comparison of Tn21 and its three derivatives.