Figures & data
Table 1 RT-PCR Primer Sequences
Figure 1 The growth of standard bacteria, susceptible and drug-resistant Escherichia coli in the culture medium.
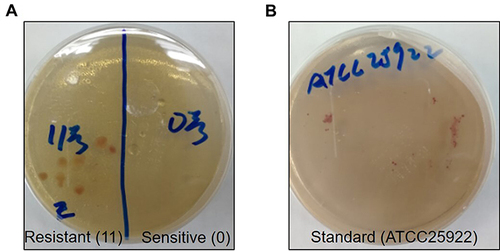
Figure 2 Sequencing results of the whole genome map of drug-resistant bacteria.
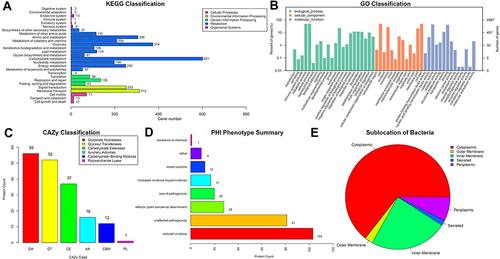
Figure 3 Relative gene expression of different types of bacteria (using 16s as the internal reference).
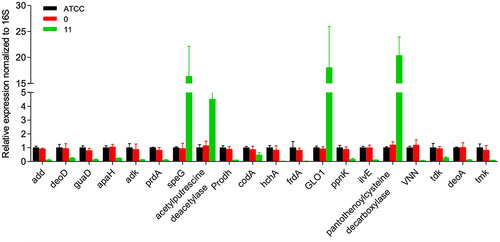
Figure 4 Verification of Glo1 gene knockout and overexpression in bacteria strain.
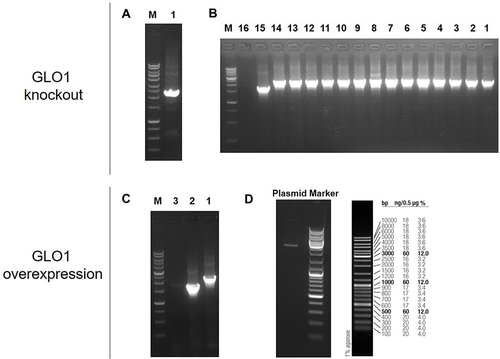
Figure 5 Determination of β-lactamase content in bacteria by ELISA.
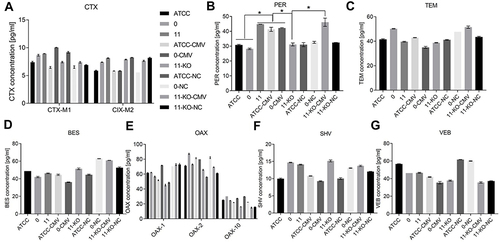
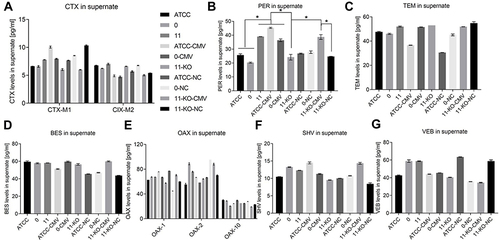
Figure 6 Determination of β-lactamase content in supernatant by ELISA.