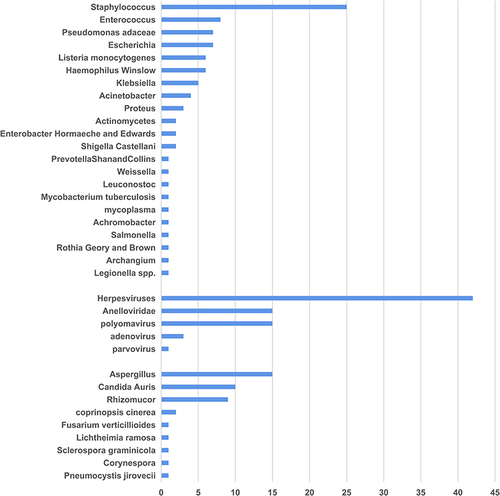
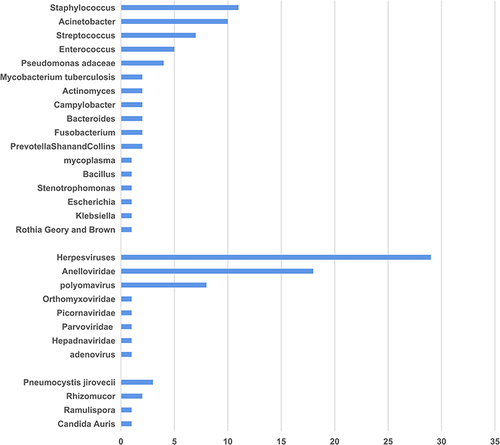
Figures & data
Table 1 Characteristics of Patients
Table 2 Detection Efficiency of mNGS and Comparison in FN and Non-FN Groups
Figure 3 The difference between mNGS and traditional method.
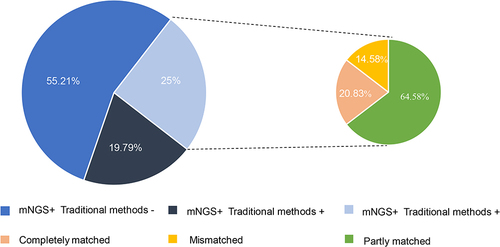
Table 3 Diagnostic Efficiency of Culture and mNGS Compared to Clinical Diagnosis
Table 4 The mNGS Results That Led Discontinued Inappropriate Antibiotics
Table 5 Diagnostic Value of mNGS in Fungal Infection in Patients with FN