Figures & data
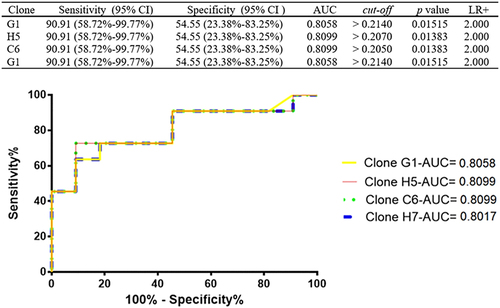
Figure 1 Comparison of the reactivity of clones identified by phage display between the sera of patients who were positive and negative for C. trachomatis. (A) Phage ELISA with the clones selected by biopanning and sequenced. The clones marked with “*” were selected for use in the ELISA optimization test. Evaluation of reactivity by phage ELISA of individual samples with clones G1 (B), H5 (C), H7 (D) and C6 (E).
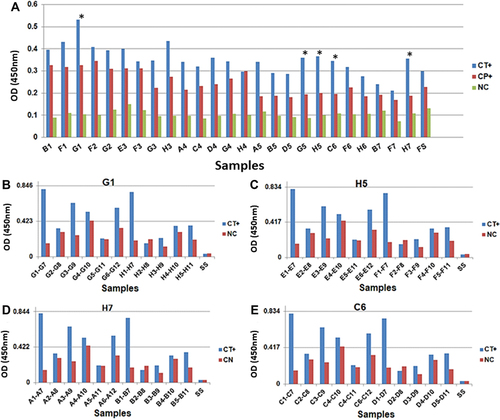
Table 1 Amino Acid Sequences of the Clones That Showed the Best Performance in the Phage ELISA Tests