Figures & data
Table 1 Basic information of 62 patients with active TB
Table 2 Basic information of healthy individuals
Table 3 Primer Sequences for Real-Time PCR
Figure 1 Diversity of gut microbiota in each patients with RR-TB, Pre-XDR, MDR-TB and healthy individuals.
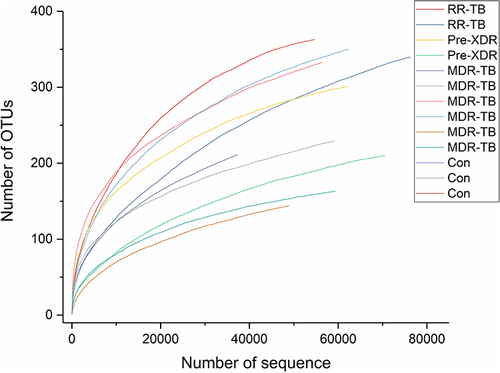
Figure 2 Variety differences of gut microbiome (different treatments vs healthy individuals).
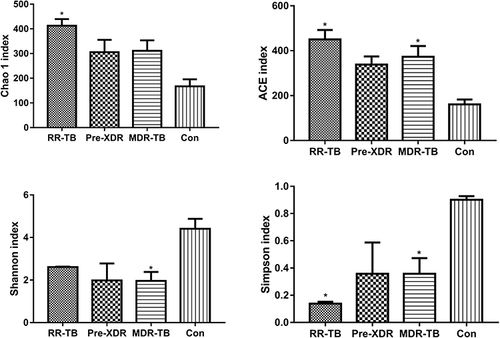
Figure 3 Distribution of gut microbiome in different subjects at the phylum level.
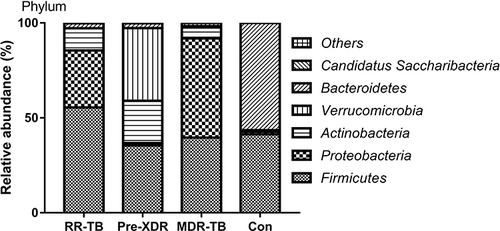
Figure 4 Distribution of gut microbiome in different subjects at the order level.
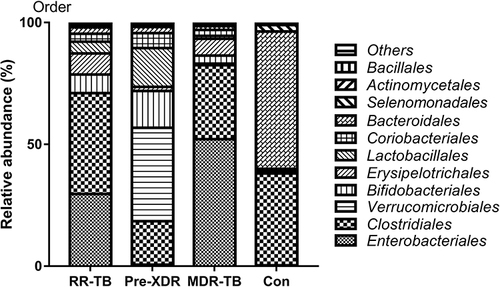
Figure 5 Distribution of gut microbiome in different subjects at the family level.
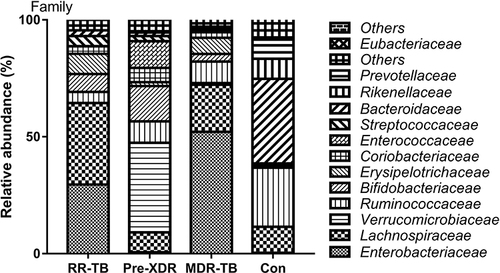
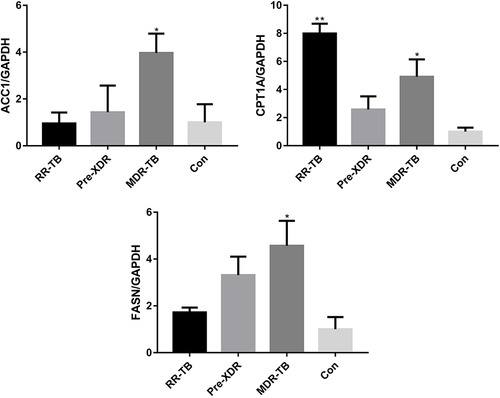
Figure 6 Differences of mRNA expression of CPT1, GAPDH, FASN and ACC1 in de novo synthesis of fatty acids.