Figures & data
Table 1 Characteristics of the Four Patients with Keratitis and Three Healthy Individuals
Table 2 Genes Validated by Quantitative RT-PCR
Figure 1 DEGs in the P. aeruginosa isolates from keratitis and healthy conjunctival sacs. Scatter plot (A) and volcano plot (B) of the upregulated and downregulated DEGs were shown. Genes with a corrected p value of less than 0.05 found with DEseq were assigned as differentially expressed.
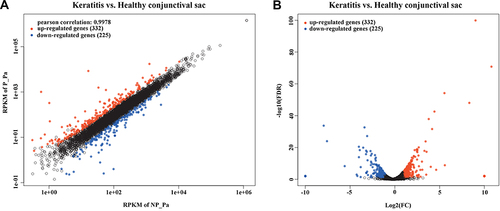
Figure 2 GO enrichment analysis of DEGs in the keratitis group compared with the healthy conjunctival sacs group. The top 30 enriched GO terms were shown. Enriched GO terms with a corrected p value of less than 0.05 were considered significantly enriched.
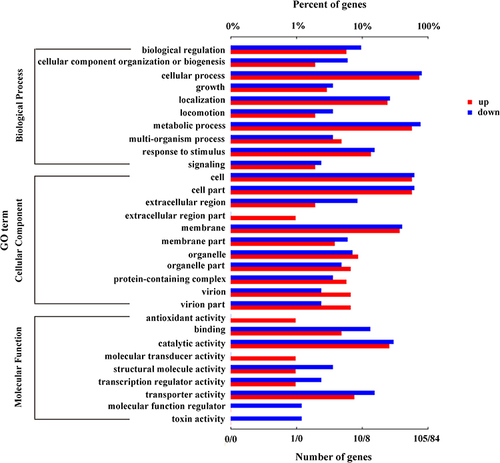
Figure 3 KEGG pathways of DEGs in the keratitis group compared with the healthy conjunctival sacs group. The top 30 upregulated (A) and the top 29 downregulated (B) KEGG pathways of the DEGs were shown. Enriched pathways with an adjusted p value of less than 0.05 were considered significantly enriched.
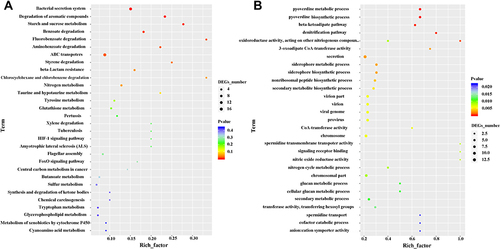
Figure 4 Validation of DEGs using real-time PCR. A total of 11 genes (log2 fold change >2 and adjusted p <0.05) from the significantly enriched KEGG pathway were chosen to be validated. The mRNA expression of pyoverdine metabolism-related genes (A), T2SS- and pili-related genes (B), and T3SS-related genes (C) were shown. Each genes were validated in all the P. aeruginosa isolates and the results showed as mean ± standard deviation of all P. aeruginosa isolates from keratitis or the healthy conjunctival sac group. (*p<0.05, **p<0.01, ***p<0.001, n = 3). (Control: healthy conjunctival sac).
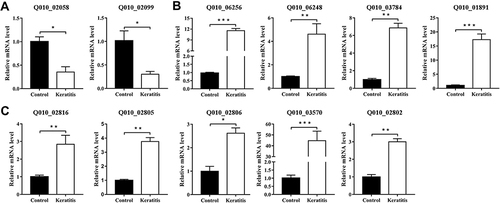
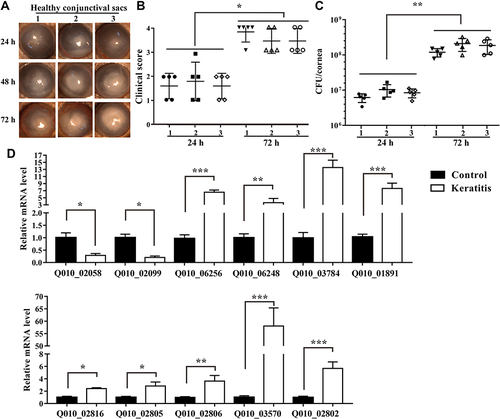
Figure 5 Changes of the DEGs of P. aeruginosa isolates from eye with healthy conjunctival sac to keratitis. Representative slit lamp microscopy images at 24, 48, 72 h (A), clinical scores (B), and bacterial load (C) at 24 and 72 h after bacterial inoculation (1, 2, 3: healthy conjunctival sac number), and qRT-PCR results of DEGs of P. aeruginosa from murine infectious keratitis at 48 h after bacterial inoculation and healthy conjunctival sac (D) are shown (*p < 0.05, **p < 0.01, ***p < 0.001, n = 5). (Control: healthy conjunctival sac).