Figures & data
Figure 1 In order to screen the mutant strains, we used flow cytometry to measure the fluorescence of GFP in mutated S. xylosus, after seven, nine, and eleven generations.
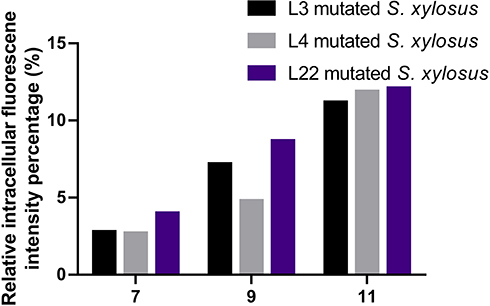
Figure 2 (A) 3D model of S. xylosus L22. The overall structure consists of three α-helices and three β-strands. The encircled box indicates the location of β-hairpin. (B) The amino acid sequence of L22 together with distribution of particular secondary structures. (C) The overall structure of S. xylosus ribosomal subunit. The 23S rRNA is shown in gray, the L22 is shown in orange, the antibiotic-binding site is shown in cyan, and the PTC location is marked as a purple dot.
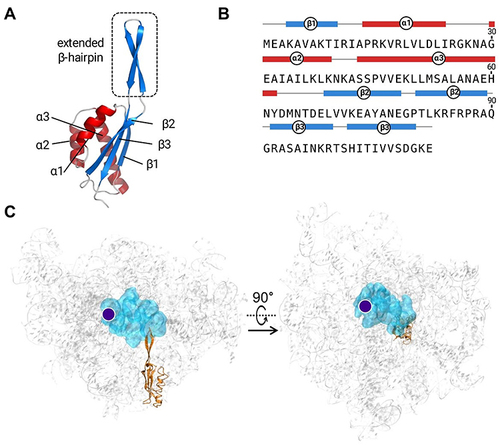
Figure 3 A model of antibiotics in complex with the S. xylosus ribosome. (A) Superposition of the binding locations of tylosin (red) and florfenicol (green). Surface representation of the structure of 23S rRNA (cyan) and L22 (orange). Tylosin binds to the peptide exit tunnel, whereas florfenicol is above the exit tunnel of PTC peptide (B). The hydrogen bond interactions (purple dash) of florfenicol (C) and tylosin (D) with 23S rRNA in the ribosome.
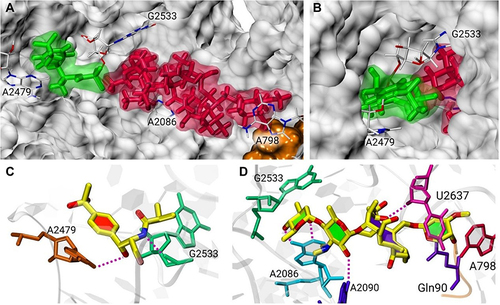
Figure 4 L22 resistance mechanism. (A) Secondary structure changes of the L22 β-hairpin during MD simulations in the wild type (upper panel) and 97KRTSAIN98 insertion mutant (lower panel). (B) Intrinsic mobility of the L22 mutant calculated from PCA analysis. The lengths of the green vectors scale with the domain motions. (C and D) Structure of the nascent polypeptide in the exit tunnel. The model of the nascent polypeptide chain is shown in red. The structure of the ribosome shown comprises the 23S rRNA and the homology model of L4 and L22 mutants. The 23S rRNA is shown in gray, the antibiotic-binding site in cyan, L4 in green, and L22 in orange. Wild-type L22 is represented as a white cartoon (D).
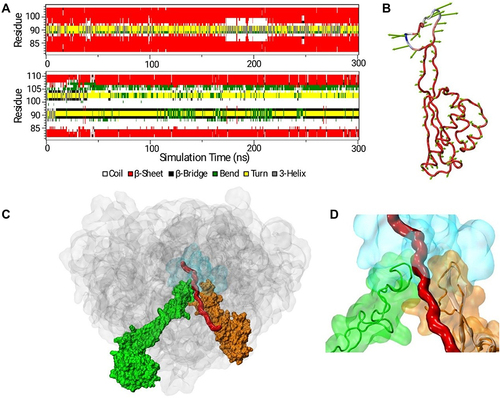
Figure 5 Changes in resistant proteins at mRNA level of different tylosin-resistance levels in S. xylosus, including thioredoxin (trxA), 50S ribosomal protein L23 (rplW), aldehyde dehydrgenase (aldA-1), chaperonin (gros), L-lactic dehydrogenase (ldh), and chloramphenicol-resistant protein (cl).
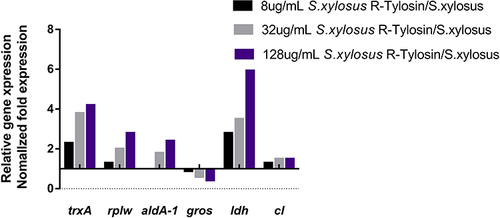
Figure 6 Changes in resistant proteins at mRNA level of L22 (rplV) mutant S. xylosus, including thioredoxin (trxA), 50S ribosomal protein L23 (rplW), aldehyde dehydrgenase (aldA-1), chaperonin (gros), L-lactic dehydrogenase (ldh), and chloramphenicol-resistant protein (cl).
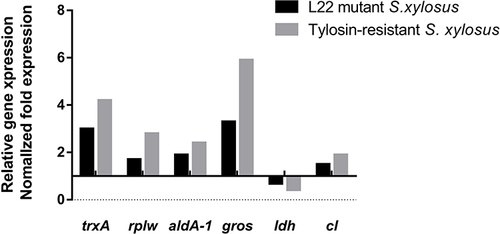
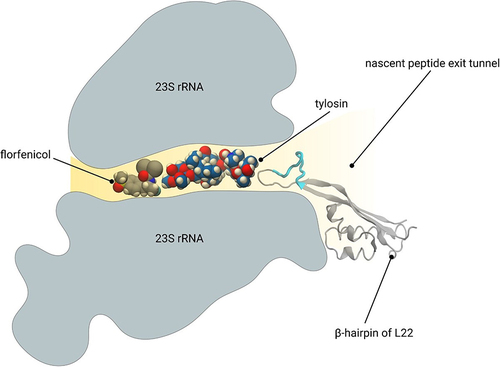
Figure 7 The drug-resistance mechanism of tylosin and florfenicol. Florfenicol (gray and red spheres) binds to the deep narrow pocket of the nascent peptide exit tunnel, which is far away from the β-hairpin of L22. A loop (in cyan) inserted into the β-hairpin of L22 has limited influence on the drug resistance of florfenicol. Tylosin (blue and red spheres), which is much larger than florfenicol, binds to the entrance of the nascent peptide exit tunnel. It closes into the β-hairpin of L22. The insertion of additional amino acid (in cyan) shows great impact on the drug resistance of tylosin.