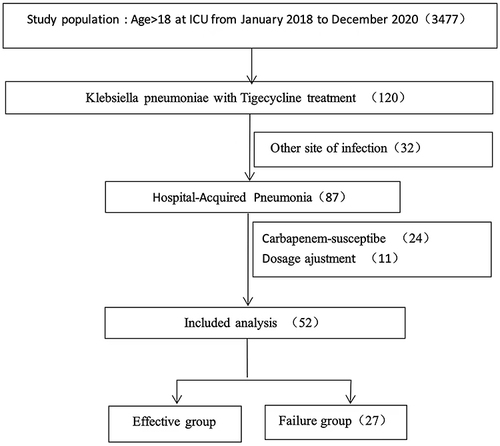
Figures & data
Table 1 Characteristics of Patients with HAP Caused by KPC with TG in Both Groups
Figure 2 Distribution map of drug resistance genes. (1) Multi-drug resistance genes were detected from studied strains, including multidrug resistance efflux pump genes- major facilitator superfamily transporter(MFS), resistance-nodulation-cell division transporter (RND), and β-lactamase genes. Among them, KPC-2, SHV, CTX-M, MFS (mdth, mdtg, mdtk) and RND (acrb, tolc) genes existed in all strains. (2) The number of tetracycline resistance genes contained in the studied strains was different, in which the detection rates of teta, tetd and tetc were 6%, 4% and 100%, respectively.
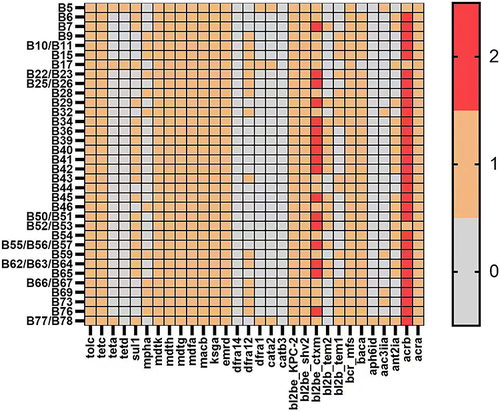
Figure 3 Distribution map of virulence genes. (1) Multiple virulence genes were detected in the studied strains, among which areo, salmonin and yersinomycin were present in all strains. (2) The detection rates of rmpA, LPS and capsular virulence genes were different among the studied strains, in which rmpA1, rmpA2, LPS-wbbo, capsular-wza and wzc were 67%, 69%, 45%, 82% and 78%, respectively.
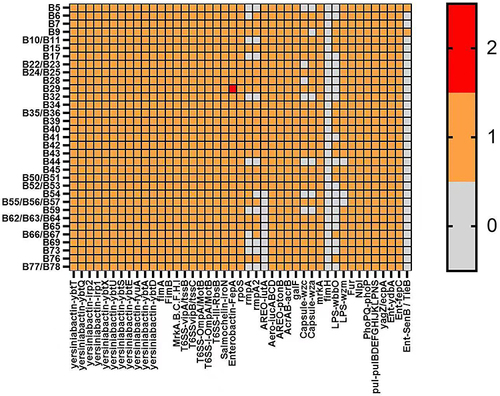
Table 2 The Mortality, Clearance of Pathogens and Distribution of Antibiotic Resistance Genes, Virulence Factors in Both Groups
Table 3 Univariate and Multivariate Analyses of the Risk Factors Associated with Clinical Efficacy in Patients with Klebsiella Pneumoniae Carbapenemase 2-Producing K. Pneumoniae Infections