Figures & data
Table 1 Genomic Information of N. terpenica NC_NT001
Table 2 Genomic Information of Nocardial Strains for Pangenomic Analysis
Figure 2 Genomic information of Nocardia terpenica NC_YFY_NT001. (A) Circular representation of N. terpenica NC_YFY_NT001 chromosome and plasmid; (B) NT001 genome-related KEGG pathway; (C) COG categories of NT001 genome (Cellular Processes and signaling: [D] Cell cycle control, cell division, chromosome partitioning; [M] Cell wall/membrane/envelope biogenesis; [N] Cell motility; [O] Post-translational modification, protein turnover, and chaperones; [T] Signal transduction mechanisms; [U] Intracellular trafficking, secretion, and vesicular transport; [V] Defense mechanisms; [W] Extracellular structures; [Y] Nuclear structure; [Z] Cytoskeleton. Information storage and processing: [A] RNA processing and modification; [B] Chromatin structure and dynamics; [J] Translation, ribosomal structure, and biogenesis; [K] Transcription; [L] Replication, recombination and repair. Metabolism: [C] Energy production and conversion; [E] Amino acid transport and metabolism; [F] Nucleotide transport and metabolism; [G] Carbohydrate transport and metabolism; [H] Coenzyme transport and metabolism; [I] Lipid transport and metabolism; [P] Inorganic ion transport and metabolism; [Q] Secondary metabolites biosynthesis, transport, and catabolism. Poorly Characterized: [R] General function prediction only; [S] Function unknown) (D) Subcellular localization of NT001.
![Figure 2 Genomic information of Nocardia terpenica NC_YFY_NT001. (A) Circular representation of N. terpenica NC_YFY_NT001 chromosome and plasmid; (B) NT001 genome-related KEGG pathway; (C) COG categories of NT001 genome (Cellular Processes and signaling: [D] Cell cycle control, cell division, chromosome partitioning; [M] Cell wall/membrane/envelope biogenesis; [N] Cell motility; [O] Post-translational modification, protein turnover, and chaperones; [T] Signal transduction mechanisms; [U] Intracellular trafficking, secretion, and vesicular transport; [V] Defense mechanisms; [W] Extracellular structures; [Y] Nuclear structure; [Z] Cytoskeleton. Information storage and processing: [A] RNA processing and modification; [B] Chromatin structure and dynamics; [J] Translation, ribosomal structure, and biogenesis; [K] Transcription; [L] Replication, recombination and repair. Metabolism: [C] Energy production and conversion; [E] Amino acid transport and metabolism; [F] Nucleotide transport and metabolism; [G] Carbohydrate transport and metabolism; [H] Coenzyme transport and metabolism; [I] Lipid transport and metabolism; [P] Inorganic ion transport and metabolism; [Q] Secondary metabolites biosynthesis, transport, and catabolism. Poorly Characterized: [R] General function prediction only; [S] Function unknown) (D) Subcellular localization of NT001.](/cms/asset/d9b27259-130f-4920-ae1f-fb7f510bdf02/didr_a_12153418_f0002_c.jpg)
Figure 3 Pan-genomic analysis of Nocardia spp. (A and B) A pangenomic curve of Nocardia spp. (C) COG analysis for pangenomic, core, shell, and cloud clusters. (D) Number of genes in pangenomic, core, shell, and cloud clusters. (E) The phylogenic tree of Nocardia spp. core genes.
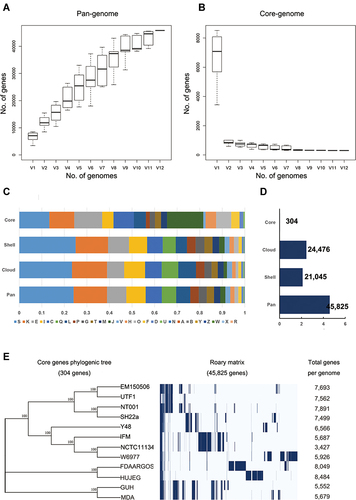
Figure 4 Comparisons of virulence-associated genes in Nocardia spp. (A) Putative virulence factors in different Nocardia spp.; (B) Toxin–antitoxin-related genes in different Nocardia spp.
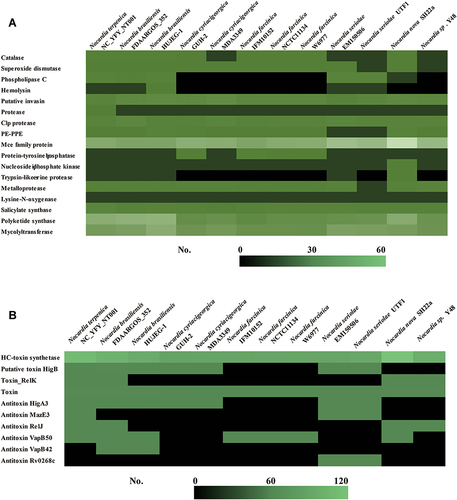
Table 3 Virulence Factors in Different Nocardial Strains