Figures & data
Table 1 Antimicrobial Susceptibility Profile of the Test CRKP Isolates
Figure 1 Antibiotic resistance profile of the test Klebsiella pneumoniae isolates. Out of 134 Klebsiella pneumoniae isolates, 35.07% and 30.6% demonstrated resistance to imipenem and meropenem, respectively. High resistance profiles were observed for most of the investigated β-lactams, including the third generation cephalosporins CTX (98.51%), CAZ (96.27%), and CRO (94.03%). Resistance rates to fluoroquinolones were recorded at 87.31% for CIP and 85.07% for LEV.
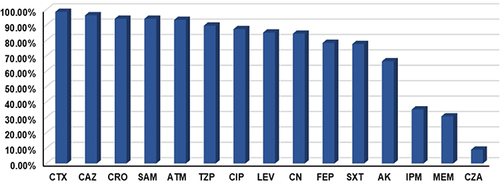
Table 2 Risk Factors and Outcome of Ceftazidime/Avibactam Resistance Amongst the Study Cohort
Table 3 Multivariate Analysis for Predictors of Ceftazidime/Avibactam Resistance Amongst the Study Cohort
Table 4 Phenotypic and Genotypic Characterization of the Investigated CRKP Isolates (N= 47)
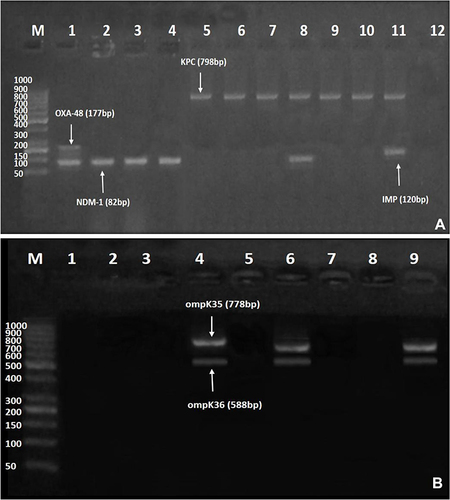
Figure 2 (A and B) Gel electrophoresis of multiplex polymerase chain reaction (PCR) amplified products for carbapenemase-encoding and outer membrane porin genes amongst ceftazidime/avibactam-resistant isolates. (A) blaNDM−1 (82 bp), blaIMP (120 bp), blaOXA-48 (177 bp), and blaKPC (798 bp). Lane M: DNA Ladder (1000 bp); Lane 1: positive result for both blaOXA-48 and blaNDM−1; Lanes 2–4: positive result for blaNDM-1; Lanes 5–10: positive result for blaKPC; Lane 8: positive result for both blaKPC and blaNDM−1; Lane 11: positive result for both blaKPC and blaIMP; and Lane 12: negative control. (B) ompK35 (778 bp), and ompK36 (588 bp). Lane M: DNA Ladder (1000 bp); Lanes 1, 2, 3, 5, and 7: negative result for both ompK35 and ompK36; Lanes 4 and 6: positive result for both ompK35, and ompK36; Lane 8: negative control for both ompK35 and ompK36; Lane 9: Klebsiella pneumoniae (ATCC 13883) positive control that harbors both ompK35, and ompK36.