Figures & data
Figure 1 (A) Scheme of the research design. A total of five groups of 8-week-old male BALB/c mice (n = 35) were continuously treated with piperacillin-tazobactam (TZP, n=7), ceftriaxone (CRO, n=7), tigecycline (TGC, n=7), levofloxacin (LEV, n=7) or normal saline (Ctrl, n=7), respectively, for up to 4 weeks. The fecal samples and serum before (0W) and after treatment for 1 and 4 weeks were used for 16S rRNA gene sequencing. Blood was before (0W) and after treatment for 1 and 4 weeks for the determination of serum cytokines. (B) Effect of antibiotic treatment on mouse growth performance. Time curve of body weight ratio changes at different time points. 0: before treatment. 3, 7, 14, 21 and 28: 3 7, 14, 21 and 28 days after treatment.
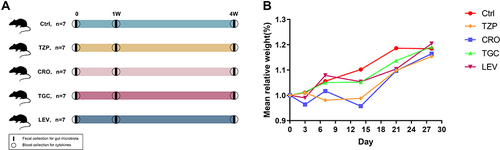
Figure 2 Changes in levels of serum cytokines after antibiotics treatment for 4 weeks. (A) IL-1α, (B) IL-1β, (C) IL-3, (D) IL-4, (E) IL-6, (F) IL-9, (G) IL-12 (p40), (H) IL-17, (I) IFN-γ, (J) eotaxin, (K) GM-CSF, (L) MCP-1, (M) MIP-1α, (N) MIP-1β, (O) RANTES, (P) TNF-α. Significant differences between the two groups were marked, and *Indicates p < 0.05; **p < 0.01 and ***p < 0.001.
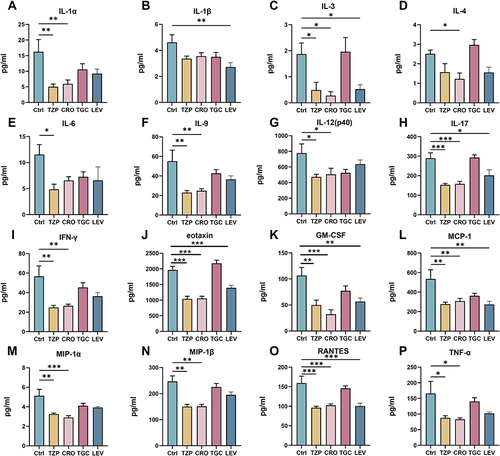
Figure 3 Microbial diversity measured by the Shannon index and Ace index was altered by antibiotic treatment across the treatment groups on 1W (A and B) and 4W (C and D). *P < 0.05, **P < 0.01 vs the control group. Analysis of microbial communities by principal coordinate analysis (PCoA) at different time points after antibiotic treatment on 1W and 4W (E and F). 1 W and 4 W: 1 W and 4W after treatment.
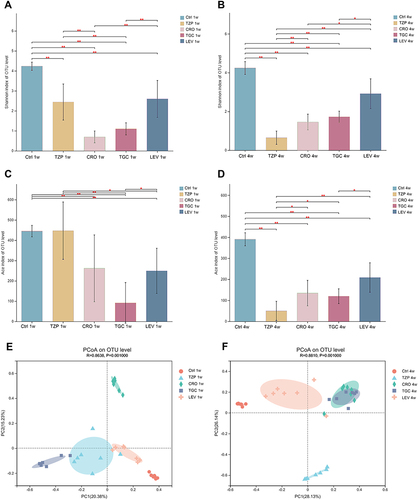
Figure 4 Antibiotic-induced composition changes of gut microbiota. Distribution of the predominant bacteria with antibiotics-treated at (A) phylum level, (B) family level and (C) genus level on 0W, 1W and 4W. Species with less than 0.5% abundance in all samples were merged into the “Others” category. 0W: before treatment. 1 W and 4 W: 1 W and 4W after treatment.
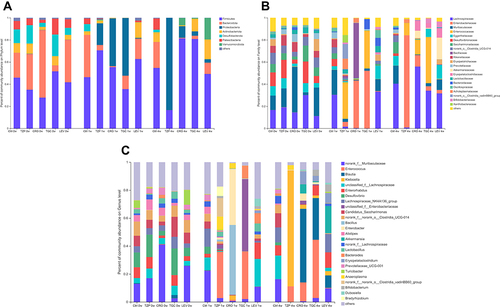
Figure 5 Comparison of dominant bacteria at the (A) Phylum, (B) Family and (C) Genus levels among five groups on 1W. Comparison of dominant bacteria at the (D) Phylum, (E) Family and (F) Genus levels among five groups on 4W. 1 W and 4 W: 1 W and 4W after treatment.
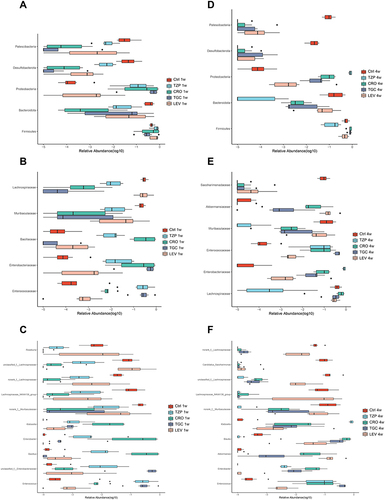
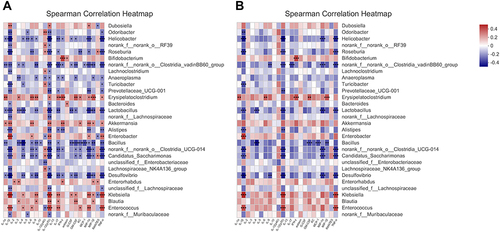
Figure 6 Correlations microbiome compositions (at the top 30 genus level) and the 23 serum cytokine levels of host immune markers on 0W, 1W and 4W. (A) Significance thresholds included absolute correlation coefficients higher than 0.1 and P < 0.05. (B) Significance thresholds included absolute correlation coefficients higher than 0.3 and P < 0.05. The number in color present Spearman correlation coefficient: red color indicates positive correlation; blue color indicates negative correlation. *P < 0.05, **P < 0.01, ***P < 0.001.