Figures & data
Figure 1 Timeline to summarize the case history of the patient admission, treatment, and isolate collection.
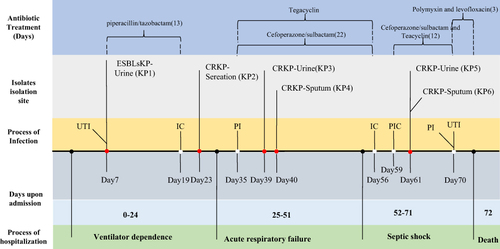
Table 1 Antimicrobial Susceptibility Test of Six Klebsiella pneumoniae Isolates
Figure 2 Determination of carbapenem resistance of the KP strains by measuring carbapenemase activity using NG-test CARBA 5 kit (A), and carbapenem resistance genes using PCR (B). C: positive control, K: KPC, O: OXA-48-like, V: VIM, I: IMP, N: NDM.
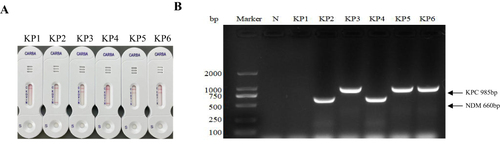
Table 2 Summary of the Genetic and Phenotypic Profiles of the Klebsiella pneumoniae Isolates
Table 3 Viable Counts (CFU/mL) and Percentage of Living Colonies in Serum Killing Assay
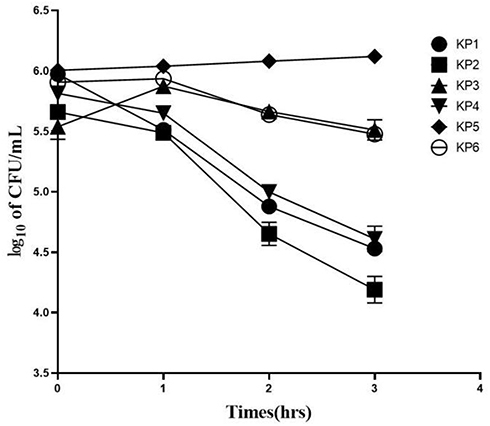
Figure 4 Logarithmic calculation of viable colony counts (CFU/mL) of the KP strains subjected to co-incubation with pooled healthy serum for three hours. The values were expressed in the form of mean ± error, and log 10 was used for logarithmic conversion to simplify the data.