Figures & data
Table 1 Sequences of Oligonucleotides Used in This Study
Table 2 Identified Accuracy Rate of Streptococci by Two MALDI-TOF Systems
Figure 1 The molecular phylogenetic tree of viridans group streptococci isolates based on tuf gene. The evolutionary history was inferred by using the Neighbor-Joining method. The optimal tree with the sum of branch length = 5.29069834 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood methodCitation2 and are in the units of the number of base substitutions per site. Evolutionary analyses were conducted in MEGA7. The length of the compared sequences was 506 bp.
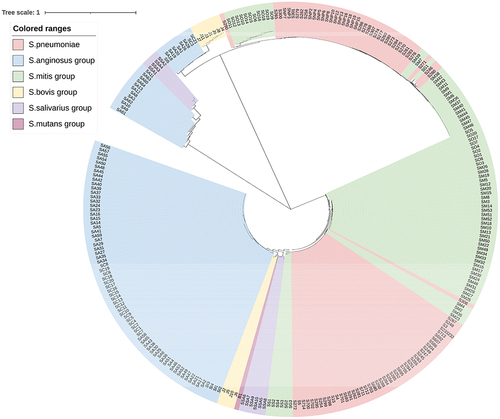
Table 3 Identification Results of MALDI-TOF Systems for Alpha-Hemolytic Streptococci
Table 4 Error Identification of Streptococci by Two MALDI-TOF MS Systems
