Figures & data
Table 1 Clinical Characteristics of 180 Patients with Pneumonia According to Immunocompromised Status and Pneumonia Severity
Figure 1 The positive rate comparison of detected microorganisms by mNGS and CMT in patients with pneumonia. (A) Comparison of positive rate between pairwise mNGS and CMT in 180 patients with pneumonia, considering results of all sample types. (B) Comparison of positive rate between pairwise mNGS and CMT in 180 patients with pneumonia, only considering CMT results of sputum samples. (C) Pie chart demonstrates the positive and negative distribution of mNGS and CMT results. mNGS+: positive only by mNGS; CMT+: positive only by CMT; double+: both positive by CMT and mNGS; double-: both negative by mNGS and CMT. (D) For the double positive subgroup (grey in pattern c), 87 patients were divided into match (12/87), partial match (62/87) and mismatch (13/87). Match: both positive by CMT and mNGS and pathogens completely overlapped; mismatch: conflicts between mNGS and CMT; partial match: both positive by CMT and mNGS and partial overlapping of microorganisms.
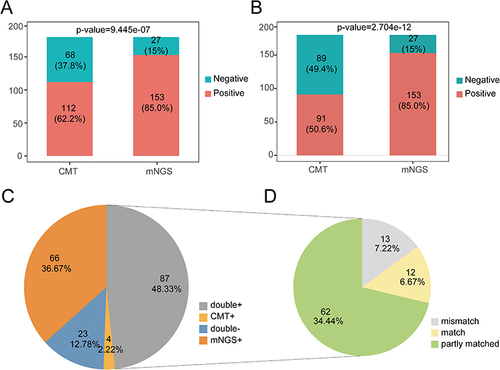
Table 2 The Concordance Between Microorganisms Detected by mNGS and the Finally Definitive Pathogens Confirmed by the Clinician
Figure 2 The overlap and comparison of positive microorganisms between mNGS and CMT in 180 patients with pneumonia. (A) Bacteria levels; (B) fungi level; and (C) virus level.
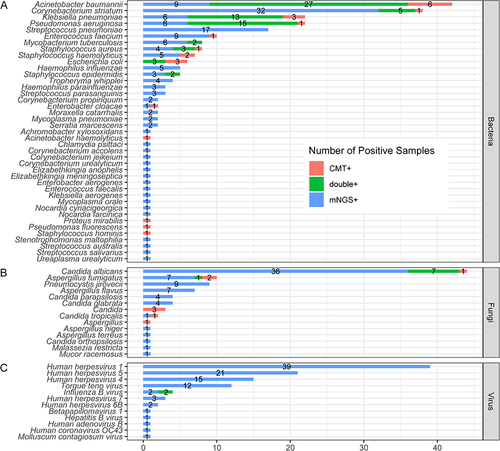
Table 3 The Concordance of Results Between mNGS and CMT Was Analyzed Using Cohen’s Kappa Statistics in Patients with Non-Severe and Severe Pneumonia
Figure 3 The positive rate comparison of detected microorganisms by mNGS and CMT between non-severe and severe pneumonia patients. (A) Comparisons of positive rates for pairwise mNGS and CMT test in non-severe and severe pneumonia patients. (B) Comparison of positive rate and mixed infection (at least two) rate by mNGS between non-severe and severe patients.
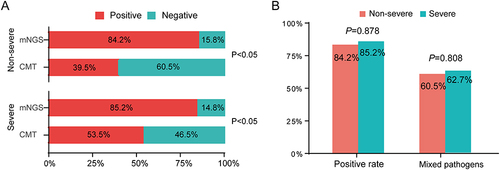
Figure 4 Comparison of microorganisms detected by mNGS between non-severe and severe pneumonia patients in bacteria (A), fungi (B) and virus (C) levels.
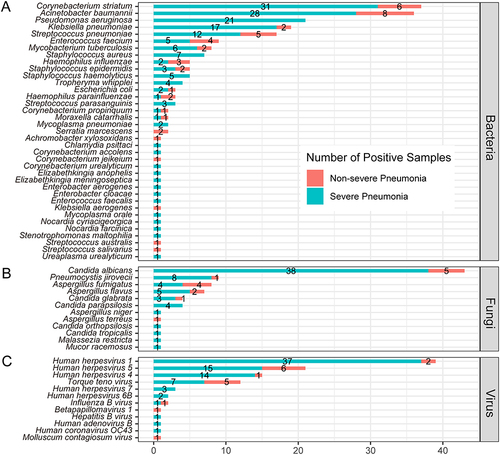
Table 4 The Concordance of Results Between mNGS and CMT Was Analyzed Using Cohen’s Kappa Statistics in Immunocompetent and Immunocompromised Patients with Pneumonia
Figure 5 The positive rate comparison of detected microorganisms by mNGS and CMT in immunocompetent or immunocompromised patients with pneumonia. (A) Comparisons of positive rates for pairwise mNGS and CMT test in immunocompetent and immunocompromised patients with pneumonia. (B) Comparison of positivity rate of mixed infection based on mNGS between immunocompetent and immunocompromised patients with pneumonia.
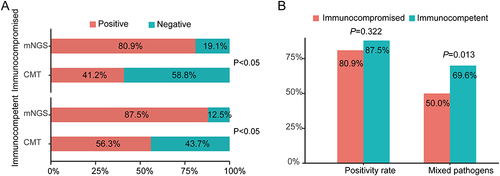
Figure 6 Comparison of microorganisms detected by mNGS between immunocompetent and immunocompromised patients with pneumonia in bacteria (A), fungi (B) and virus (C) levels.
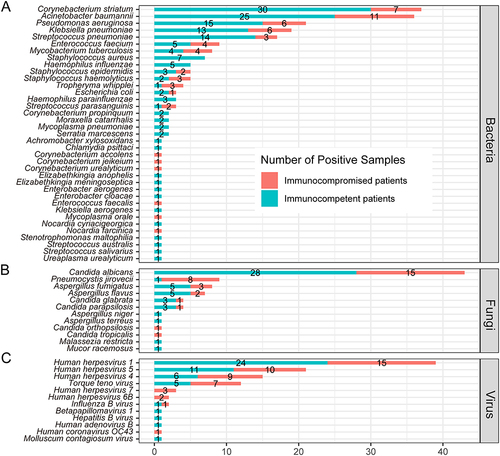
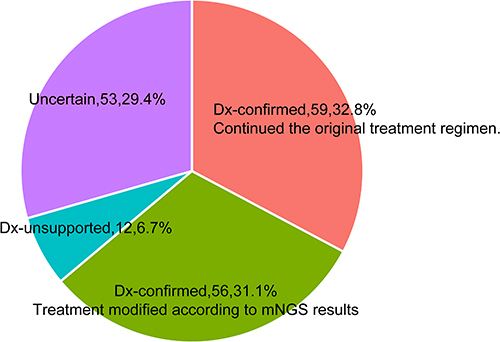
Figure 7 The concordance between microorganisms detected by mNGS and the finally definitive pathogens confirmed by the clinicians. Dx-confirmed: the microorganisms detected by mNGS were finally confirmed as pathogens of pneumonia by clinicians; Dx-unsupported: the microorganisms detected by mNGS were not confirmed as pathogens of pneumonia by clinician. Dx-uncertain: the pathogen of pneumonia was unproven.