Figures & data
Table 1 Sequencing and qRT-PCR primers
Figure 1 Positions of overlapping primers on the crtOPQMN operon.
Note: The green triangles indicate the position of overlapping primers used for the amplification and sequencing of the crtOPQMN operon.
Abbreviations: crtOPQMN, carotenoid biosynthetic operon; F, forward; R, reverse; crtO, glycosyl-4,4′-diaponeurosporenoate acyltransferase; crtI, phytoene dehydrogenase; crtQ, 4,4′-diaponeurosporenoate glycosyltransferase; crtM, dehydrosqualene synthase; crtN, dehydrosqualene desaturase.

Figure 2 The effect of co-culture with different bacteria on pigment production by the Staphylococcus aureus white variant.
Notes: The co-culture experiment was carried out on an LB agar plate for 48 hours between the Staphylococcus aureus white variant (W) and Escherichia coli Mg1655, Acinetobacter baumani/co 41-00-sc1-c1, Enterobacter W001, or Pseudomonas aeruginosa 6611. Only P. aeruginosa 6611 induced pigment production in the white variant (yellow colony).
Abbreviation: LB, luria-Bertani.
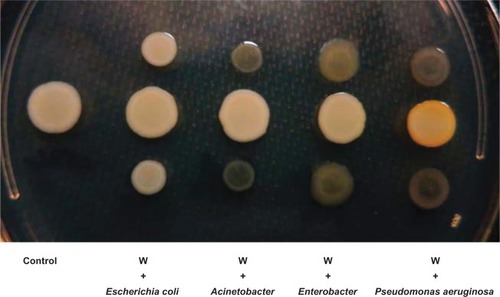
Figure 3 HPTLC analysis of staphyloxanthin in crude methanol extract from Staphylococcus aureus. Crude methanol extracts of staphyloxanthin from the S. aureus white variant (W), the S. aureus white variant co-cultured for 48 hours with Pseudomonas aeruginosa 6611 (Wp), and the S. aureus yellow variant (Y) were analyzed by HPTLC on silica plates (pore size 60 Å). The results indicated a significant difference in pigment-band intensity between the white variant and the white variant co-cultured with P. aeruginosa 6611.
Notes: Numbers below the bands represent the absorbance of the crude methanol extracts at A465 (W = 0.121, Wp = 0.463, Y = 0.381). There is ~4-fold increase in absorbance in crude extracts of the white variant after co-culture with P. aeruginosa 6611, indicating substantial increase in pigment production.
Abbreviation: HPTLC, high performance thin layer liquid chromatography.
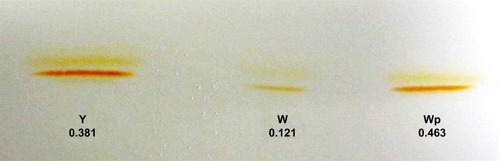
Figure 4 Transcriptional analysis of selected genes from the Staphylococcus aureus white variant after co-culture with Pseudomonas aeruginosa. Several genes involved in pigment production were analyzed by qRT-PCR after co-culture with P. aeruginosa 6611. These genes included crtM, which is directly involved in staphyloxanthin synthesis. Transcriptional factors SigB and Sara control the expression of the crtOPQMN operon, and product of the hfq gene may be involved in post-transcriptional control of the operon. FPP-synthase gene is involved in the synthesis of farnesyl-diphosphate which is the substrate for the crtOPQMN operon. The results indicate no significant changes in expression of these genes through the first 7 hours of co-culture. In the later time intervals of 24 hours and 48 hours, these genes (with the exception of sarA which was slightly downregulated) were significantly downregulated. The agrA gene could be involved in the downregulation of staphyloxanthin. The expression of the katA gene may be correlated to the production of staphyloxanthin since both confer resistance to H2O2. The transcription of agrA is downregulated after 7 hours and into the later time intervals. In contrast, katA gene is significantly upregulated after 8 hours and reached its peak (more than 3-fold) at 24 hours.
Notes: The columns represent fold change in gene expression of co-cultured S. aureus white variant as compared to the white variant that was not co-cultured. Time points represent data gathered after 1, 7, 24, and 48 hours after the start of co-culture with P. aeruginosa 6611.
Abbreviations: qRT-PCR, quantitative reverse transcription polymerase chain reaction; crtM, carotenoid biosynthetic gene M; sigB, sigma B transcription factor gene; sarA, staphylococcal accessory regulator A gene; FPP synth, farnesyl pyrophosphate synthase gene; hfq, host factor gene for Q beta; katA, catalase gene A; agrA, accessory gene regulator A; crtOPQMN, carotenoid biosynthetic operon.
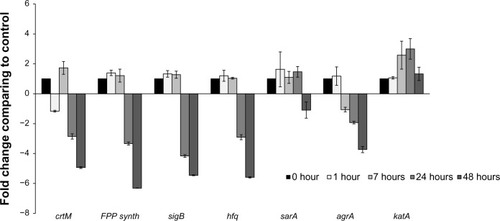
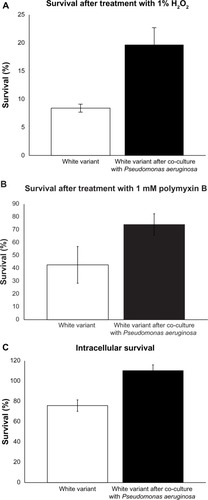
Figure 5 Induction of resistance to stress in the Staphylococcus aureus white variant by Pseudomonas aeruginosa 6611. (A) co-culture with P. aeruginosa 6611 for 48 hours led to enhanced survival of the white variant in the presence of 1% H2O2 as compared to the white variant without co-culturing (21.8% ± 3.1% vs 8.9% ± 0.7%, P < 0.05). Induction of staphyloxanthin production and upregulation of catalase gene could account for increased resistance of the white variant to H2O2. (B) co-culture with P. aeruginosa 6611 for 48 hours increased the resistance of the white variant to 1 mM polymyxin B. The difference between survival of the white variant after co-culturing and the white variant without co-culturing is almost 2-fold (75.3% ± 8.3% vs 42.7% ± 14.9%). (C) co-culture with P. aeruginosa 6611 for 48 hours resulted in enhanced survival of the white variant within the mouse macrophage cell line J7741.a. after 2 hours of infection, the number of co-cultured white variant increased slightly (110.4% ± 4.1%). In the parallel experiment with the white variant without co-culturing, the number of survived bacteria decreased after 2 hours of infection (75.9% ± 3.9%, P < 0.05).
Notes: All results represent the mean from either two experiments (panel A) or three experiments (panels B and C), expressed as a percentage of the initially inoculated number of the S. aureus white variant (white bars) and the pigmented white variant after 48 hours of co-culture with P. aeruginosa 6611 (black bars). The P-value was determined using student’s t-test.