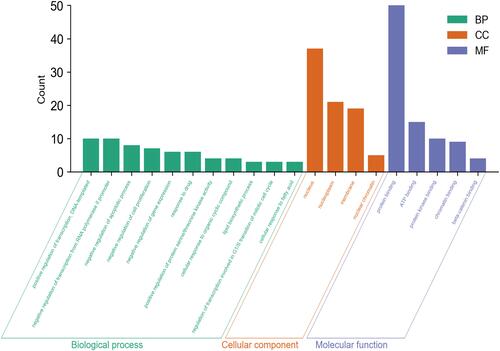
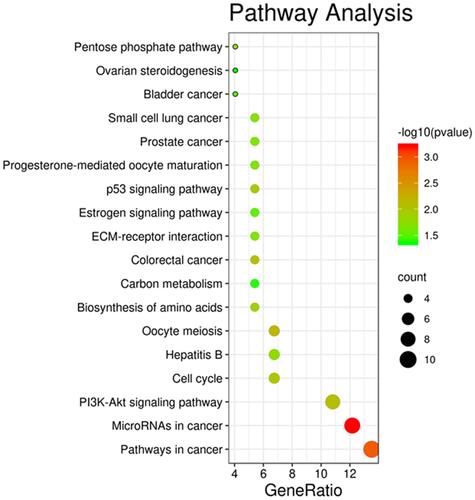
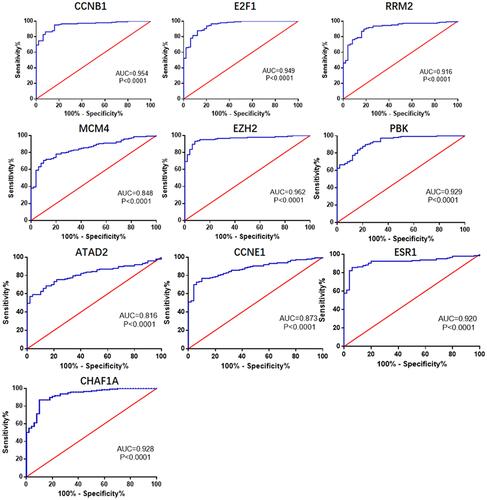
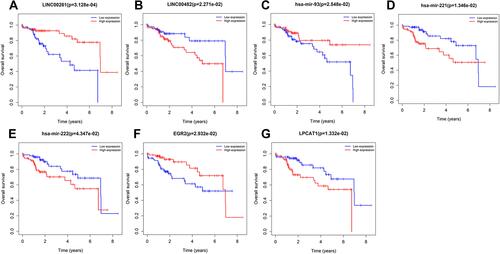
Figures & data
Figure 1 Volcano plots of differentially expressed lncRNA (A), miRNA (B) and mRNA (C) in AFP-negative HCC. Red dots indicate genes significantly upregulated and green indicates genes significantly downregulated.
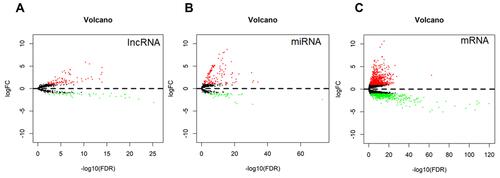
Table 1 12 DElncRNAs Target 23 DEmiRNAs from the miRcode Database
Figure 2 Venn diagram analysis for the intersection set between differentially expressed mRNA in AFP-negative HCC and miRNA target genes.
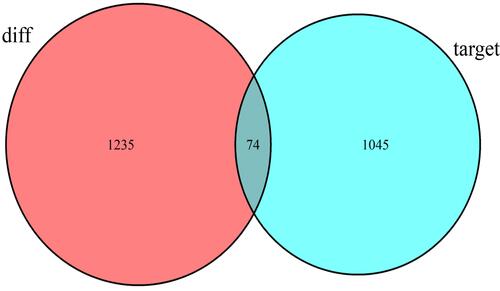
Figure 3 The lncRNA-miRNA-mRNA ceRNA network in AFP-negative HCC. The diamond represents lncRNA, the square represents miRNA and the circle denotes mRNA. The red nodes indicate upregulated RNAs and the green nodes indicate downregulated RNAs.
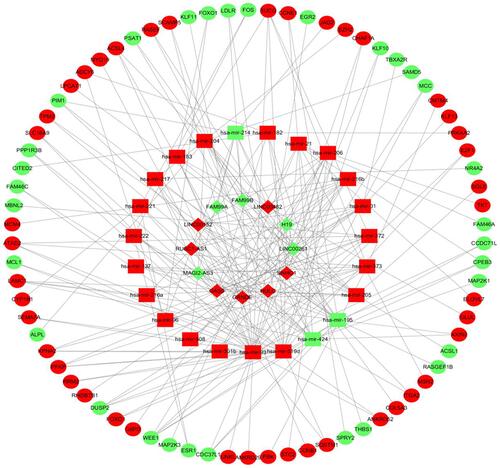
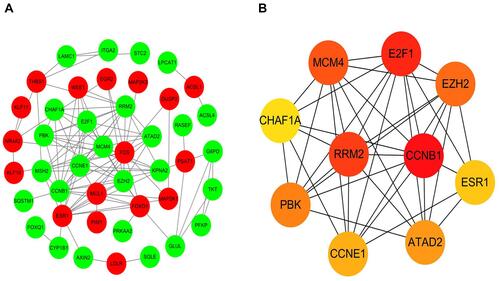
Figure 6 (A) A PPI network of DEmRNAs involved in the ceRNA network of AFP-negative HCC. Red nodes represent the upregulated DEmRNAs and green nodes represent the downregulated DEmRNAs. (B) The top ten highly interacted hub genes were recognized by cytoHubba.