Figures & data
Figure 1 Study flowchart.
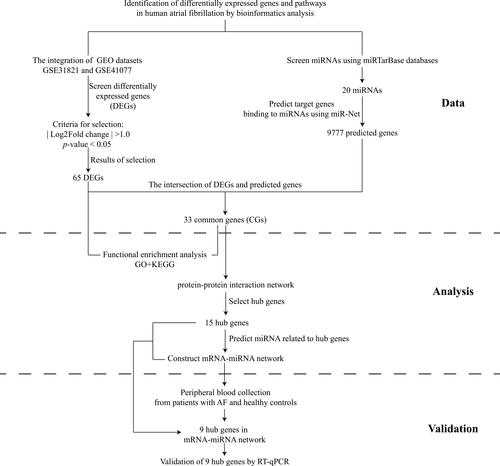
Figure 2 Volcano plots and Heatmap for the DEGs identified from the integrated dataset.
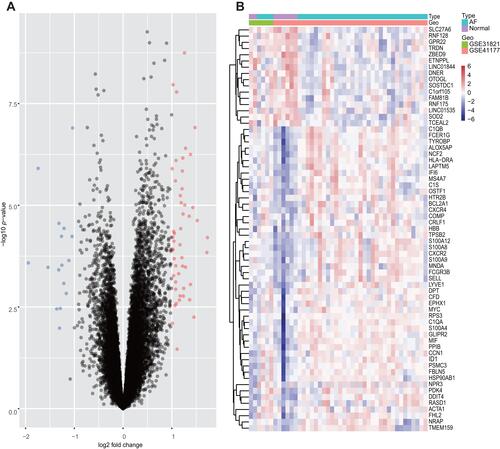
Figure 3 Terms of BP, CC and MF in pathway-enrichment analyses for DEGs using the GO database.
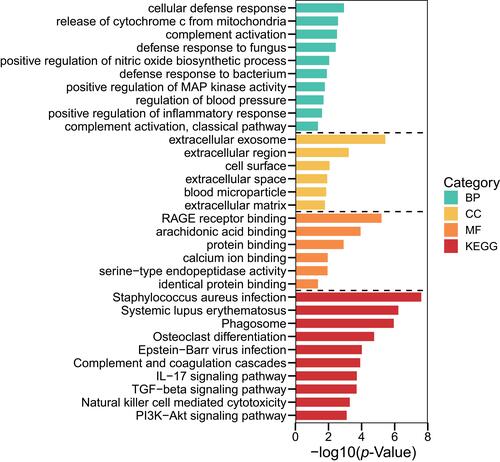
Figure 4 Venn diagram, PPI network and the functional terms and pathways enriched for common genes.
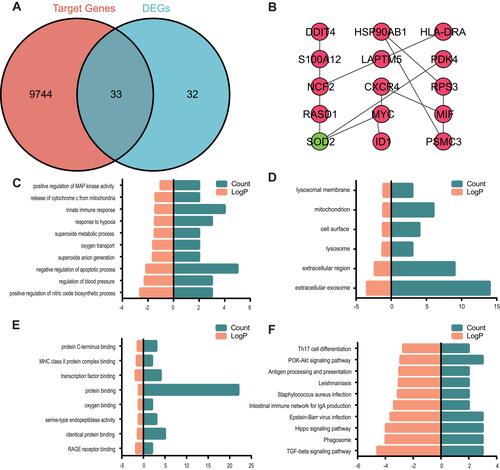
Table 1 The Nodes with Degree of ≥ 2 in the mRNA-miRNA Network
Figure 5 miRNA–mRNA integrated network.
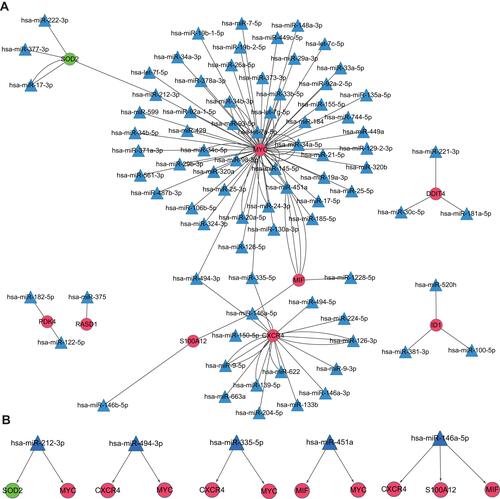
Figure 6 The drug-gene network.
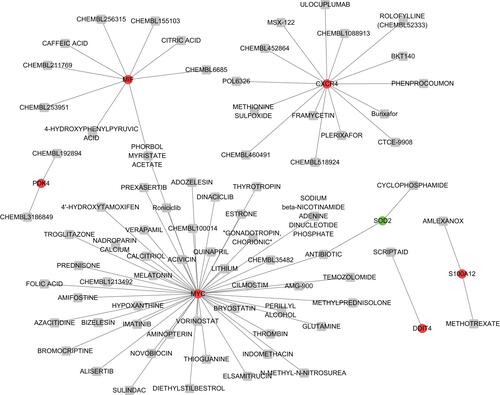
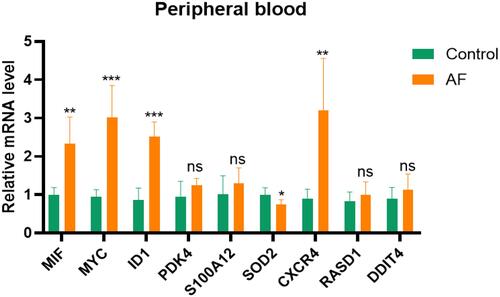
Figure 7 Validation of hub genes in AF patients.