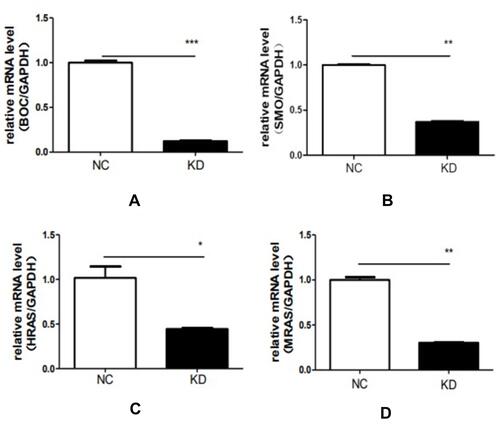
Figures & data
Table 1 Sample Grouping Schedule of GBM Tissue and Corresponding Paracancerous Enrolled in This Study
Figure 1 H&E staining result.
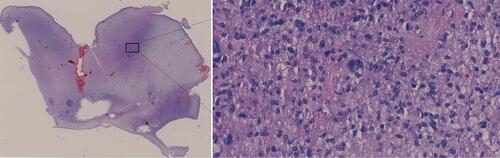
Table 2 Summary of Small RNA (sRNA) Sequencing Datasets
Table 3 List of Differentially Expressed microRNAs (DE miRNAs) in Response to GBM
Table 4 Summary of mRNA Sequencing Datasets