Figures & data
Table 1 Some Gene Signatures for Hepatocellular Carcinoma Extracted from Literature
Figure 1 A workflow diagram for the analysis. The LIMORE dataset was to build a reference network and identify modules. The preservation of identified modules was validated in two independent datasets LCCL and CCLE. These modules were used as references, to which new datasets can be projected. The derived module eigengene (ME) can be used in differential module identification, survival analysis, patient classification, and in silico drug-resistant analysis.
Table 2 Functional Annotation and Hub Gene of the 13 Identified Modules in Hepatocellular Carcinoma Cell Line
Figure 2 Gene co-expression modules identified in LIMORE HCC cell line dataset. (A) Scale-free topology model fit R2 versus power selection using FPKM; (B) scale-free topology model fit R2 versus power selection using rank; (C) the log-log plot shows an R2 of 0.95 when power set at 12, indicating the network follows the scale-free topology criterion; (D) mean connectivity versus power selection, showing the connectivity is stable at power 12; (E) thirteen modules were identified according to the dendrogram produced by hierarchical clustering of LIMORE genes based on a topological overlap matrix (TOM). The modules were assigned colors as indicated in the horizontal bar; (F) multidimensional scaling plots in two dimensions (color-coded as in E) depict the relative size of the modules.
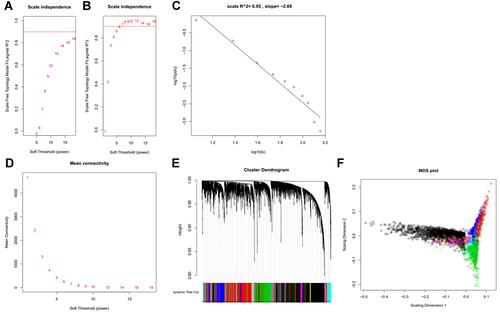
Figure 3 The identified thirteen modules are reproducible. (A) Bar plots showing the intramodule connectivity correlation of each module by half-sampling 1000 times with the original one (mean ±SD); (B) preservation of modules in a network constructed by FPKM compared to that constructed by rank. (C) preservation of modules in CCLE network compared to that in LIMORE; (D) preservation of modules in LCCL network compared to that in LIMORE. Dashed green and blue lines represent the Zsummary threshold for strong (Z>10) and weak–moderate (2<Z<10) module preservation. Numbers along with colored dots represent the identified modules.
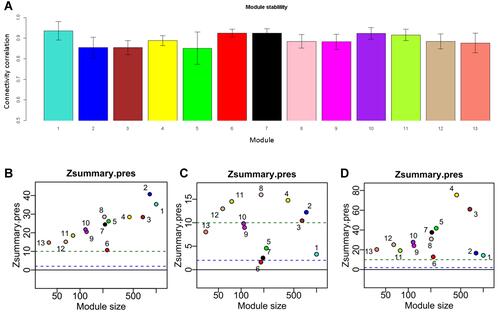
Figure 4 Differentially expressed modules (DEM) in HCC development. (A) Clustering analysis of the thirteen modules in HCC development. In the left sample bar, green, blue, yellow, and red denotes normal, HCV, HCC_cirrhosis, and HCC samples; (B) Box plot showing the M3 expression; (C) Box plot showing the M6 expression; (D) Box plot showing the M9 expression; (E) clustering analysis of the thirteen modules in normal, adjacent and HCC tissues. In the left sample bar, green, yellow and red denotes normal, adjacent, and HCC samples.
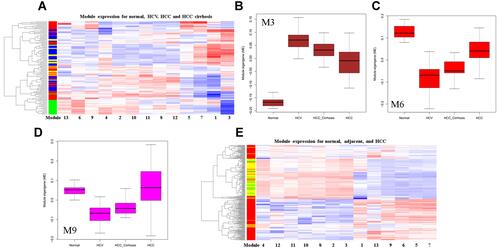
Figure 5 Box plot showing the differentially expressed modules M4 (A), M5 (B), M7 (C), and M9 (D) in malignant hepatocytes compared to non-malignant hepatocytes in the human liver.
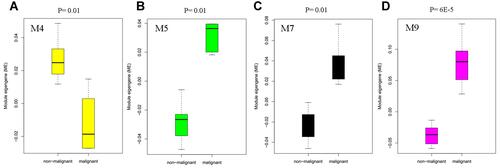
Figure 6 Modules and hub genes are indicative of patient survival. (A) Survival curves indicate that M4 and M5 expression can separate patients into two groups with different overall survival times; (B) survival curves indicate that TPX2, LUC7L3, KIF11, and modules M4, M5, M7 expression can separate patients into two groups with different disease-specific survival times; (C) survival curves indicate that ITGA3, TPX2, LUC7L3, KIF11, and modules M4, M5, M7 expression can separate patients into two groups with different disease-free survival times. Redline: low expression of genes or modules. Greenline: high expression of genes or modules.
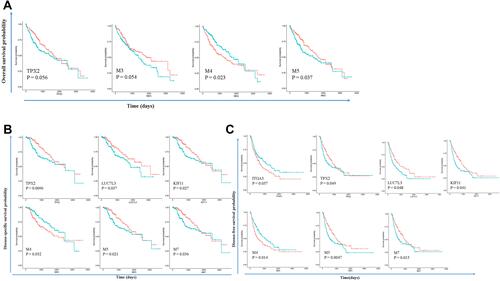
Figure 7 Top 100 connections in modules M1, M3, M5, and M7 were visualized in Cytoscape (v3.7.0). Degree analysis shows that LUC7L3, ITGA3, TPX2, and KIF11 are the hub genes in the modules. These hub genes are yellow nodes in the networks.
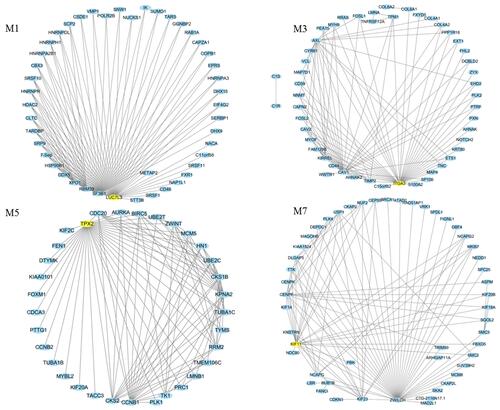
Figure 8 Representative images for immunohistochemical staining of the hub genes TPX2 (M5), LUC7L3 (M1), and KIF11 (M7), which were all up-regulated in liver cancer according to the Human Protein Atlas. The blue bar in the upper right denotes the cases with high, moderate, low, and not detected signals.
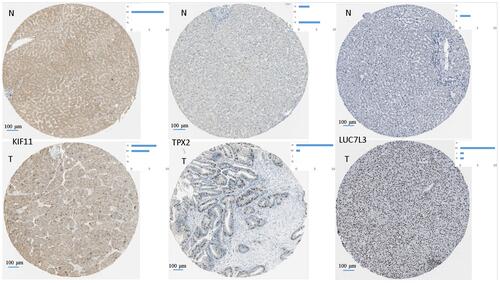
Table 3 Sensitivity, Specificity, and Area Under ROC Curve (AUC)
Figure 9 Modules M3, M5, M6, M7, and M9 have good performance in distinguishing normal liver, HCV, and HCC.
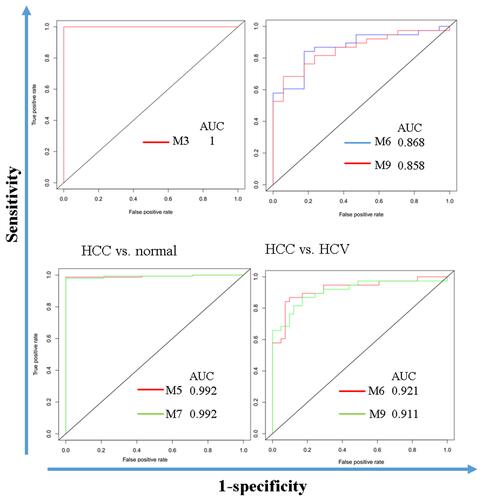
Figure 10 Correlations between module expression and IC50 reveal potential drug mechanisms of action and drug-resistant lines. (A) Workflow for drug-module association analysis; (B) drug-module correlation matrix for LCCL; (C) drug-module correlation matrix for LIMORE.
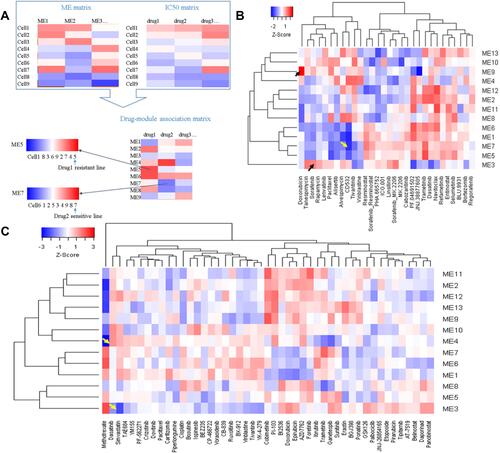
Table 4 Connectivity Map Analysis Identified 6 Candidate Drugs
