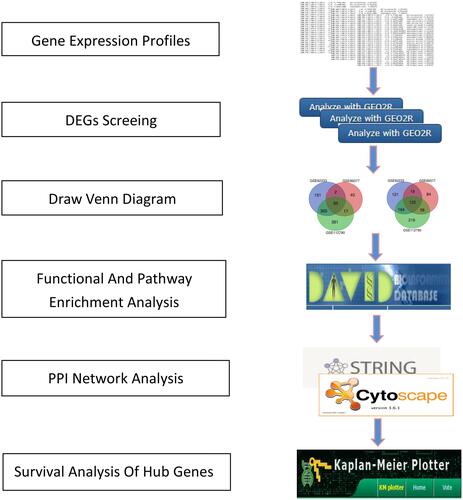
Figures & data

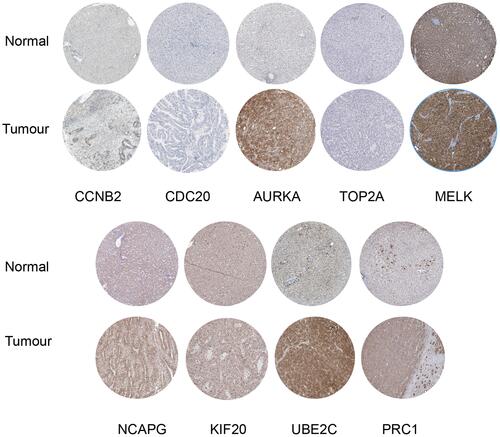
Table 1 Top Ten Hub Genes with Higher Degree of Connectivity
Table 2 Statistics of the Three Microarray Databases Derived from the GEO Database
Figure 2 Venn diagram of DEGs common to all three GEO datasets.
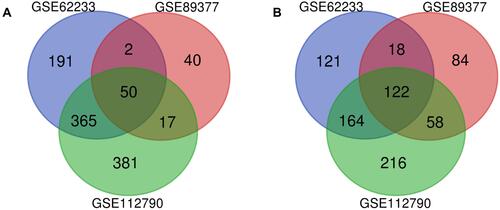
Table 3 Significantly Enriched GO Terms and KEGG Pathways of DEGs
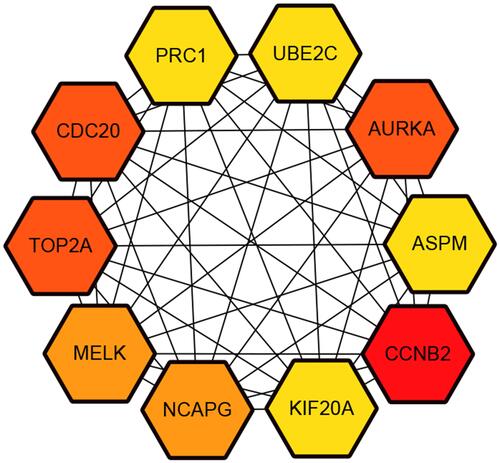
Figure 3 Protein–protein interaction network of the differentially expressed genes in hepatocellular carcinoma.
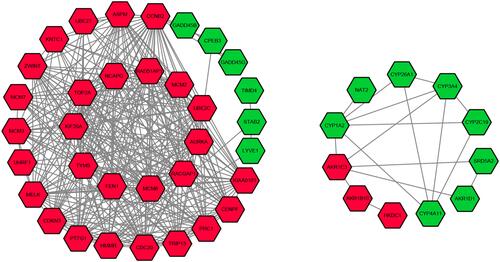
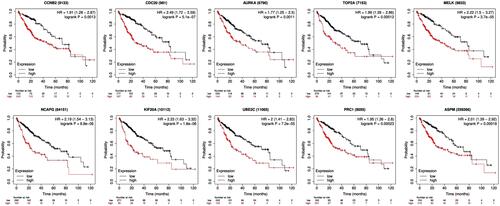
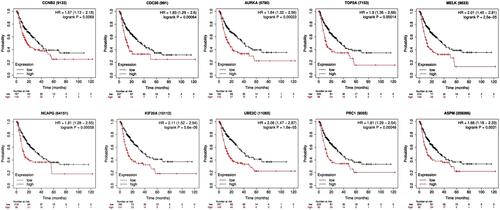
Figure 5 Kaplan–Meier overall survival analyses for the top ten hub genes expressed in hepatocellular carcinoma patients.