Figures & data

Figure 1 Association between IGFBP1 and clinicopathological characteristics of Non-small cell lung cancer (NSCLC). (A) The landscape of IGFBP1-related clinicopathological features of NSCLC in the Cancer Genome Atlas (TCGA) database. (B–E), There was no significant difference in IGFBP1 expression between NSCLC and paracancerous tissues, and between Lung adenocarcinoma (LUAD) and paracancerous tissues (Control). The expression level of IGFBP1 in lung squamous cell carcinoma (LUSC) Control and LUAD was higher than that in LUSC. (F–H), There was no difference in IGFBP1 expression levels between males and females. The expression level of IGFBP1 was relatively higher in the death group and the advanced pathological stage.
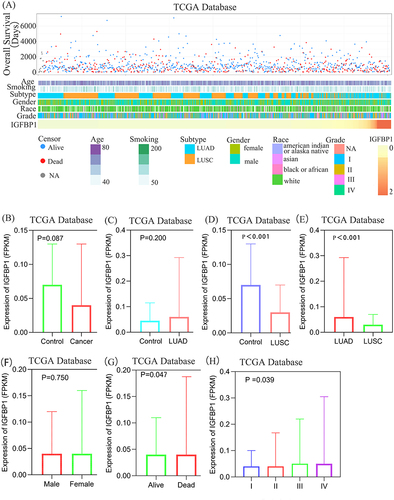
Figure 2 IGFBP1 expression is affected by common gene mutations and has a low methylation level. (A and B), Tumor type: Lung Adenocarcinoma (LUAD), Lung Squamous Carcinoma (LUSC). Mutation type: All somatic mutations, Mutation prevalence at least: 2%. P -value cutoff: 0.01. Fold change (FC) cutoff: 1.44. (C),The level of IGFBP1 promoter methylation was compared between LUAD and normal samples.(D), Differences in IGFBP1 promoter methylation levels between normal tissues and pathological stages in LUAD.(E),The level of IGFBP1 promoter methylation was compared between LUSC and normal tissues.(F), Differences in IGFBP1 promoter methylation levels between normal tissues and pathological stages in LUSC.
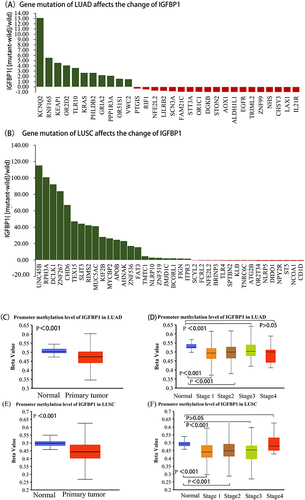
Figure 3 Gene ontology (GO) enrichment and proteome interaction analysis were performed on IGFBP1-related genes. (A–C), The biological processes (BP) accomplished by IGFPBP1 and other molecular activities were mainly the number of genes enriched most in Response to drug and cell proliferation. Regarding cellular components (CC) the Extracellular space in which IGFPBP1 gene products may reside when performing their functions. The activity of the IGFBP1 gene product complex at Molecular Function (MF) is the largest number of proteins associated with Protein binding. Negative regulation (NR), Positive regulation (PR). (D) The thickness of the lines of connection between the spheres represents the credibility of the evidence for interaction between the proteins.
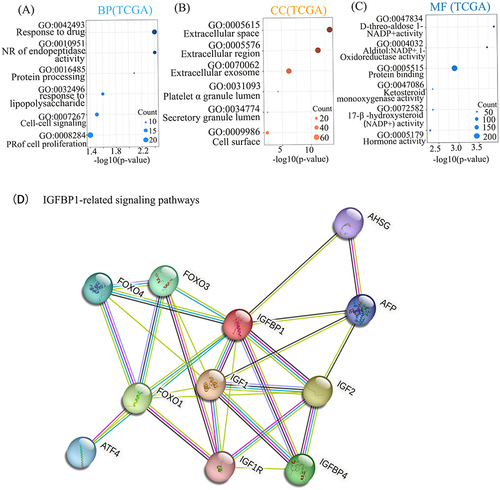
Figure 4 Kaplan-Meier analysis of IGFPB1 expression in The Cancer Genome Atlas (TCGA) databases and Kaplan-Meier Plotter. (A and B), The significance of the prognostic value was tested by a Log rank test.
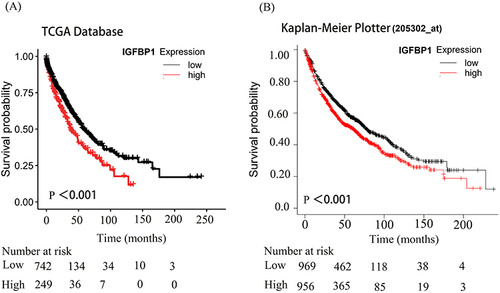
Figure 5 Timer2 was used to explore the link between IGFBP1 expression and immune infiltration. (A–D), IGFBP1 expression was positively correlated with (Myeloid-derived suppressor cells) MDSC cell infiltration and negatively correlated with Mast cell, T cell NK and B cell infiltration in lung adenocarcinoma. (E–I), Endothelial cell. In lung squamous cell carcinoma, IGFBP1 expression showed a weak positive correlation with Endothelial cells, but not with MDSC, Mast cells, T cells, NK cells and B cells. (J), IGFBP1 expression was positively correlated with SLC27A2.
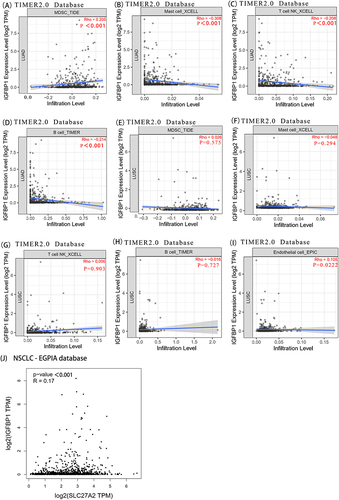
Figure 6 IGFBP1 is associated with ferroptosis inhibitors and is associated with prognosis. (A) There were 409 genes related to IGFBP1 expression. The cut-off value of correlation coefficient R was 0.2 and the cut-off value of P was less than 0.05. (B–H), The significance of the prognostic value was tested by a Log rank test. Glutamate--cysteine ligase catalytic subunit (GCS). Aldo-keto reductase family 1 member C1-3 (AKR1C1-3). Cystine/glutamate transporter (SLC7A11). Furin paired basic amino acid cleaving enzyme (FURIN). Voltage-dependent anion-selective channel protein 2 (VDAC2).
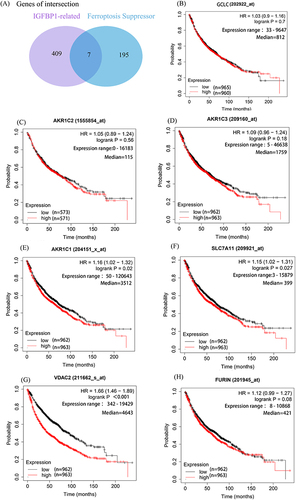
Figure 7 (A) Gene effect score for IGFBPI in Lung cancer cell lines. The Gene effect score cutoff value was 0.15. Negative scores imply cell growth inhibition and/or death following gene knockout. Scores are normalized such that nonessential genes have a median score of 0 and independently identified common essentials have a median score of −1. (B) Immunophenotyping of NSCLC. (C) Serum proteomic analysis. There are 83 similarities in proteins that can be found between the two groups. (D) Volcano plot to show significant differences between two groups of sample data. The abscissa is the fold difference (logarithmic transformation with base 2), and the ordinate is the significance P-value of the difference (logarithmic transformation with base 10). (E and F), Intersection of serum proteomics and PPI networks.
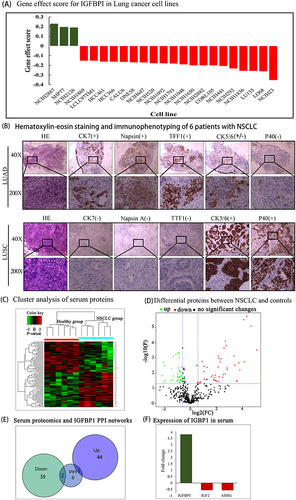
Table 1 Subject Characteristics and Protein Extraction Parameters
Table 2 Differentially Expressed Upregulated Proteins in Non-Small Cell Lung Cancer Samples
Table 3 Differentially Expressed Downregulated Proteins in Non-Small Cell Lung Cancer Samples
