Figures & data
Figure 1 DEGs were obtained by differential analysis of the eutopic (EU) and ectopic (EC) endometrium samples from the GSE141549 dataset. (A) Principal component analysis: red represents the EC group, and blue represents the EU group. (B) Volcano plot: gray denotes non-differentially expressed genes, while red and blue stand for up- and down-regulated DEGs, respectively.
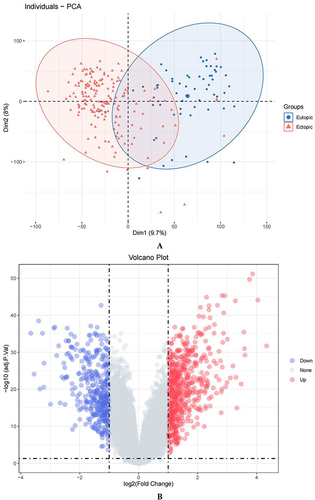
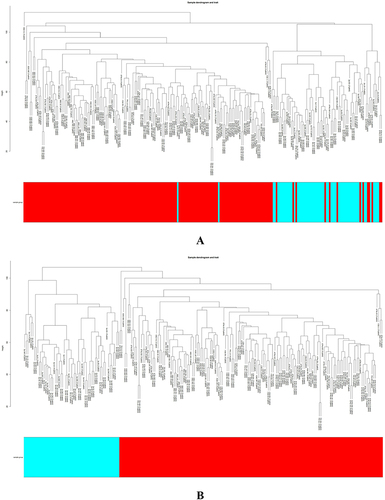
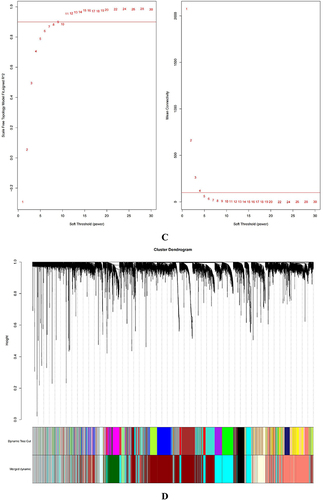
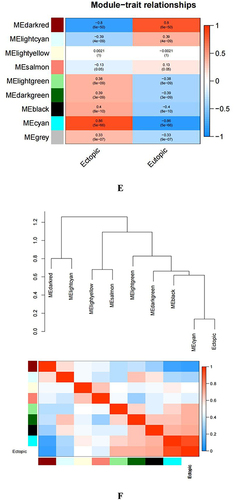
Figure 2 Identifying the essential modules linked to endometriosis tissues. (A and B) Sample cluster tree: the outlying samples are removed through the clustering book, blue represents EU, red represents EC; the upper half is a cluster, and the lower half is phenotype. (C) Screening for appropriate soft thresholds, the scale-free topology fit index value was 0.90, as indicated by the red line. (D) Dendrograms of genes with different similarity based on module colors of topological overlap and assignments. (E) Heat map of the correlation between modules and clinical characteristics. (F) Correlation of modules and EC endometrium. (G) Scatterplots of gene significance (GS) vs module membership (MM) in the cyan module.
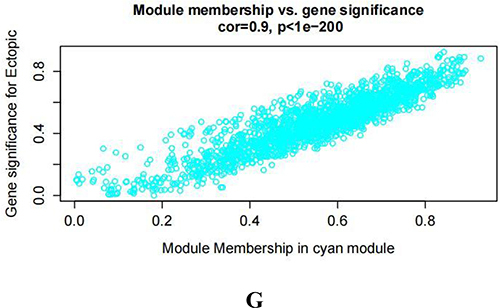
Figure 3 Screening of autoimmune gene-related genes for key module genes involved in the development of endometriosis.
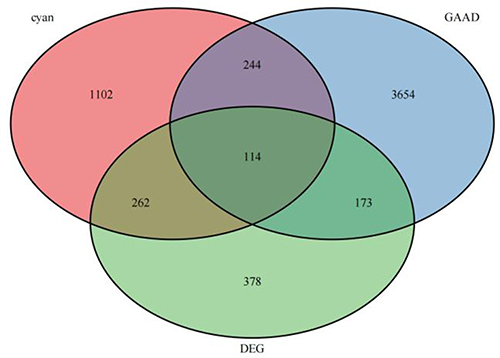
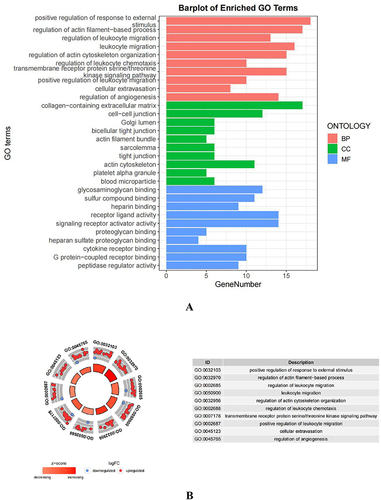
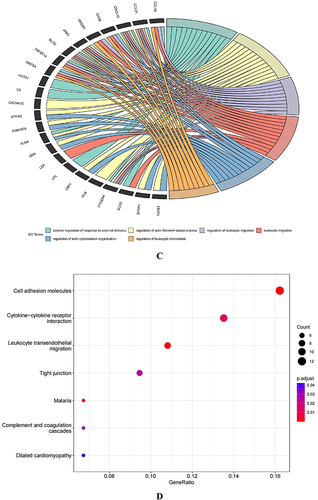
Figure 4 Functional enrichment analysis was performed for the AID-DEGs. (A) GO results of the analysis of AID-DEGs to show the enrichment results of BP, CC, and MF. (B) Circle diagram: showing the top 10 AID-DEGs. (C) The top 6 biological process GO terms and their assigned genes. (D–E) Results of the KEGG enrichment analysis of the AID-DEGs.
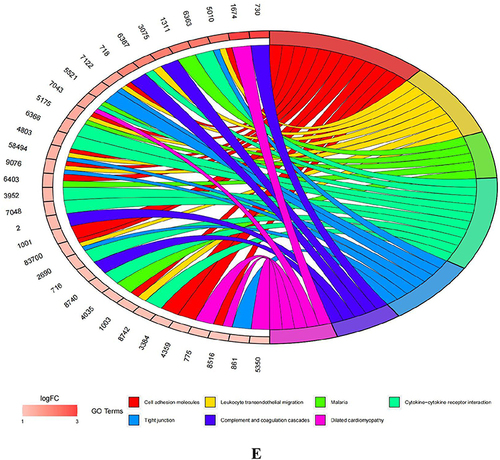
Table 1 Top 10 Essential Genes for 5 Algorithms (MCC, MNC, Degree, EPC, and Betweenness)
Figure 5 Hub genes of AID-DEGs were selected. (A) Protein interaction analysis of 114 genes using the STRING online website. (B) Venn diagram intersection of the top 10 genes.
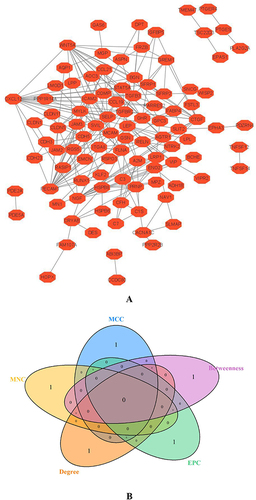
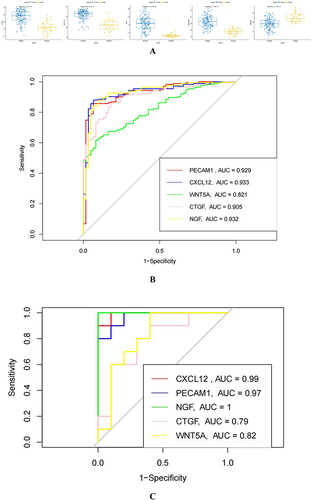
Figure 6 Analysis of differential expression and diagnostic value of hub genes in EU and EC. (A): Analysis of CTGF, CXCL12, NGF, PECAM1, and WNT5A in EU and EC. (B): At GSE141549, the diagnostic value of CTGF, CXCL12, NGF, PECAM1, WNT5A in EU and EC were analyzed. (C):The diagnostic value of CTGF, CXCL12, NGF, PECAM1, and and WNT5A was verified using the GSE7305 dataset. (D): To verify the expression difference of CTGF, CXCL12, NGF, PECAM1, and WNT5A, using the GSE7305 dataset. (E): Explore the difference in the importance of hub genes for the diagnosis of EU and EC by using the random forest algorithm. (F): Value of joint diagnosis of hub genes using SVM algorithm.
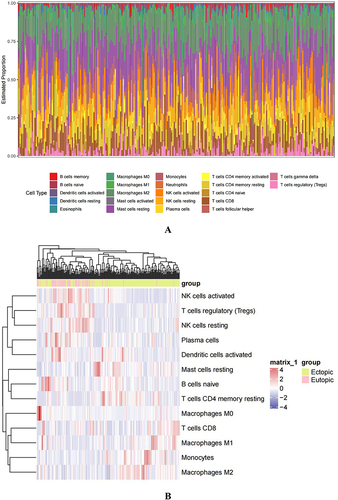
Figure 7 The association of hub genes with infiltrating immune cells. (A) Display the 22 immune cells parts for each sample. (B and C) Heatmap, box plots showing the differential infiltration of immune cells between EU and EC groups. (“*” represents P<0.05, “***” represents P<0.001, “****” represents P<0.0001) (D) Lollipop chart displaying the relationship between immune cells and hub genes.
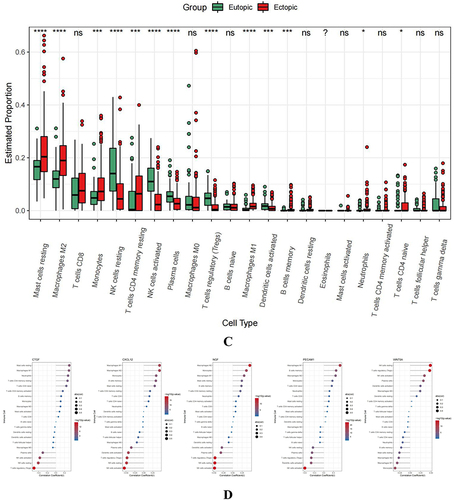
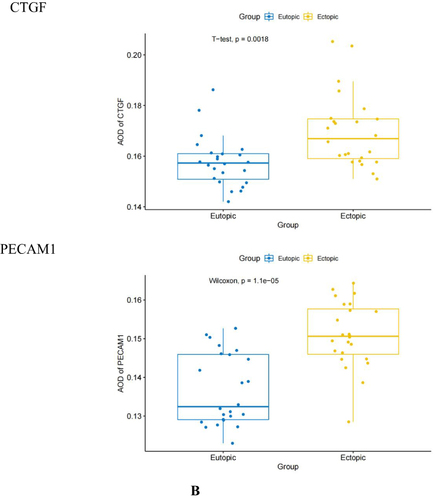
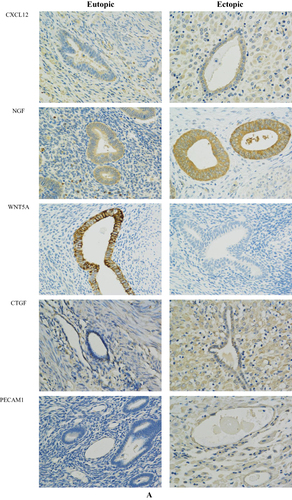
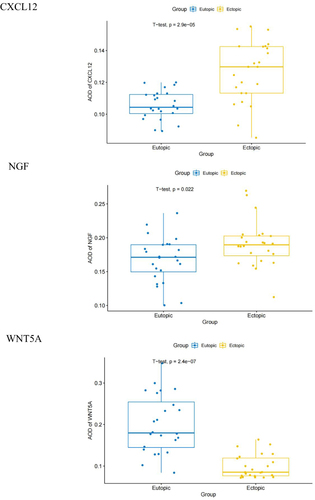
Figure 8 Immunohistochemical staining results of the EU and EC endometrium. (A) Immunohistochemical staining results of CXCL12, NGF, WNT 5 A, CTGF and PECAM1 at 400 × magnification in the EU and EC endometrium. (B) Difference in the average optical density (AOD) values of CXCL12, NGF, WNT5A, CTGF, and PECAM1 in the EU and EC endometrium.