Figures & data
Figure 1 FACS analysis of chitosan/DPP-IVODN polyplexes uptake in HepG2 cell line. Uptake of 5′-6FAM labeled DPP-IVODN in chitosanase treated and untreated cells 24 hours post transfection. (A) Transfection efficiency was calculated as the percentage of 5′FAM-DPP-IVODN labelled cells. (B) The relative amount of internalized DPP-IVODN was determined using flow cytometry and is expressed as the median of fluorescence intensity (FI) of the 6FAM-DPP-IVODN positive cells. Data points represent the mean values ± SD, n = 3.*P < 0.05; **P < 0.01.
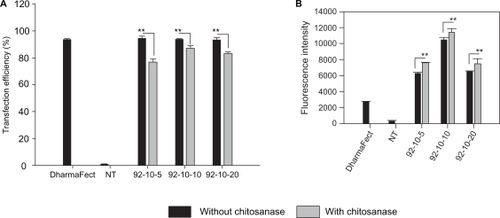
Figure 2 Confocal imaging of polyplexes uptake. Confocal microscopy images of HepG2 live cells 24 hours post transfection with chitosan/DPP-IVODN polyplexes (N/P = 5). Chitosan 92–10 (DDA, MW) was labeled with Rhodamine (red), the DPP-IVODN with 6FAM at the 5′extremity (green), and the cell membranes were stained prior to imaging with cell mask (blue). Membrane staining was performed to differentiate between internalized and membrane bound polyplexes.
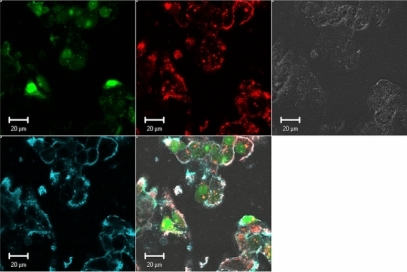
Figure 3 Effect of Streptomyces griseus chitosanase on yield and integrity of total RNA extraction. Total RNA extraction was performed on HepG2 cells transfected with 10 pmol and 50 pmol of nanoparticles siRNA/chitosan at three different N/P ratios indicated by the formulation code 92-10-5, 92-10-10, or 92-10-20 (DDA, MW, N/P). Chitosanase was resuspended in DMEM pH 6.5 and directly applied to cells at a final concentration of 6.12 mU/μg of chitosan. Total RNA was extracted from chitosan transfected cell treated with or without chitosanase. The different extractions were compared to control Dharmafect™ 1 transfected cells and nontransfected (NT) cells. (nt) = nucleotide, L = standard ladder, the green band is a lower marker, which allows sample alignment and permits comparison for RIN calculation. RIN = RNA integrity number, is an algorithm-based numbering system that calculates-RNA integrity with 10 being the most intact and 1 being fully degraded.
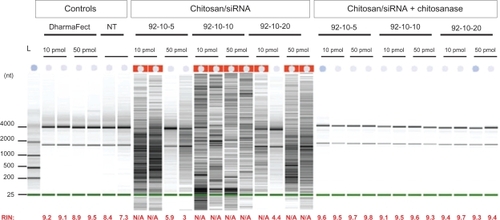
Figure 4 Polyacrylamide gel electrophoresis of chitosan/DPP-IVODN polyplexes bearing different DDAs and N/P ratios, treated with or without Streptomyces griseus chitosanase. a) chitosan migration. b) ODN migration. Lanes 1 to 4 correspond to chitosan/DPP-IVODN directly incubated with chitosanase for 60 minutes at 37°C. Chitosan digestion allows the ODN release. Lanes 5 to 8 correspond to chitosan/DPP-IVODN incubated at the similar conditions without chitosanase. Faster chitosan migration was observed when comparing lanes 5 and 6 due to different MW of theses formulations. Increased band intensity (lanes 6–8) results from greater amounts of chitosan at higher N/P ratios.
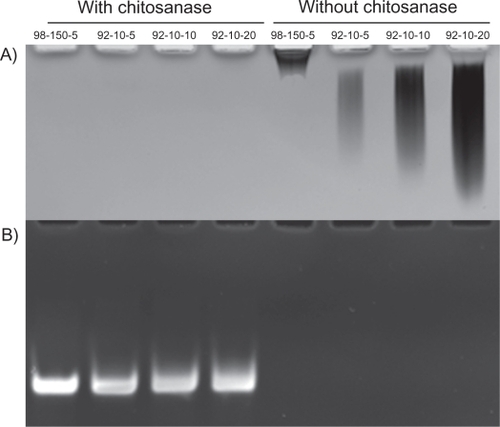
Figure 5 Total RNA extraction from HepG2 transfected cells with 10 pmol siRNA. Following transfection, cells were treated with chitosanase for: 30 min and 60 min in cell medium and for 60 min in lysis buffer.

Table 1 Effect of chitosanase treatment on RNA extraction, Relative Integrity Number (RIN), and real-time PCR (qPCR) analysis in three different DPP-IV expressing cell lines. Inhibition percentages of DPP-IV gene expression in siRNA/polyplexes transfected cells were determined in comparison with nontransfected cells