Figures & data
Figure 1 Characterization and stability study of the NLCs.
Notes: Hydrodynamic radii of PMAGP-GEM/PTX NAG-NLCs (A) and blank PMAGP NAG-NLCs (B) estimated by DLS; typical TEM images of PMAGP-GEM/PTX NAG-NLCs (C) and blank PMAGP NAG-NLCs (D). (E) Stability studies of PMAGP-GEM/PTX NAG-NLCs in double-distilled water, PBS, and 10% FBS (in PBS). Data are expressed as mean ± SD (n=3).
Abbreviations: NLCs, nanostructured lipid carriers; PMAGP, poly(6-O-methacryloyl-d-galactopyranose); GEM, gemcitabine; PTX, paclitaxel; NAG, N-acetyl-d-glucosamine; DLS, dynamic light scattering; TEM, transmission electron microscopy; PBS, phosphate-buffered saline; FBS, fetal bovine serum; SD, standard deviation.
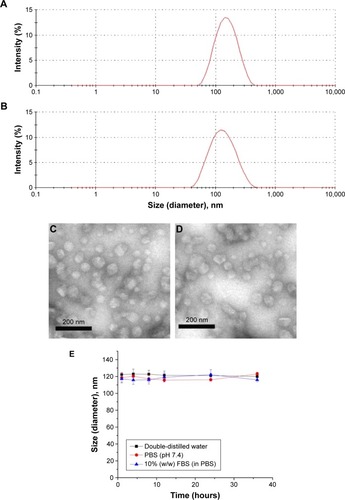
Table 1 Characteristic features of the dual drugs in PMAGP-GEM/PTX NAG-NLCs
Figure 2 The biocompatibility study of empty NLCs and in vitro cytotoxicity assay of different formulations.
Notes: (A) Cell viability of different cell types after treated with empty NLCs in vitro. A549, L929, NCIH1299, and LTEPa2 were incubated with blank PMAGP NAG-NLCs for 72 hours before MTT assay (n=6). (B) In vitro cytotoxicity of six molar ratios (5:1, 3:1, 2:1, 1:1, 1:2, and 1:3) of free GEM and free PTX formulations against A549 cells for 72 hours. (C) In vitro cytotoxicity of six molar ratios (5:1, 3:1, 2:1, 1:1, 1:2, and 1:3) of PMAGP-GEM/PTX NAG-NLCs against A549 cells for 72 hours. In vitro cytotoxicity against A549 cells (D), NCLH1299 (E), and LTEPa2 (F) for 72 hours: free GEM:PTX (3:1)-loaded NAG-NLCs (a), PMAGP-GEM NAG-NLCs + PMAGP-PTX NAG-NLCs (combo nanoparticles) (3:1)-loaded NAG-NLCs (b), PMAGP-GEM/PTX (3:1) conjugate-loaded NAG-NLCs (c), and PMAGP-GEM/PTX (3:1) conjugate-loaded NLCs (d). Data are presented as mean ± SD (n=6). **P<0.01; ***P<0.001.
Abbreviations: NLCs, nanostructured lipid carriers; PMAGP, poly(6-O-methacryloyl-d-galactopyranose); NAG, N-acetyl-d-glucosamine; GEM, gemcitabine; PTX, paclitaxel; SD, standard deviation.
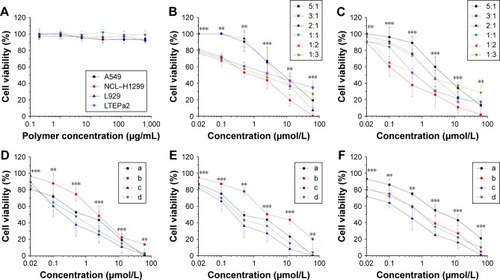
Table 2 IC50 and CI50 of different formulations in A549 cells
Figure 3 In vitro cytotoxicity assay against normal cells and in vitro drug release study.
Notes: (A) Cytotoxicity of PMAGP-GEM/PTX (3:1) NAG-NLCs and free GEM:PTX (3:1) against normal cell line L929 cells for 72 hours. Data are presented as mean ± SD (n=6). (B) Drug release profiles of PTX and GEM from PMAGP-GEM/PTX (3:1) conjugate-loaded NAG-NLCs at pH 6 and pH 7.4 in PBS containing 0.1% (w/v) Tween 80 at 37°C. Data are presented as mean ± SD (n=3).
Abbreviations: PMAGP, poly(6-O-methacryloyl-d-galactopyranose); GEM, gemcitabine; PTX, paclitaxel; NAG, N-acetyl-d-glucosamine; NLCs, nanostructured lipid carriers; SD, standard deviation; PBS, phosphate-buffered saline.
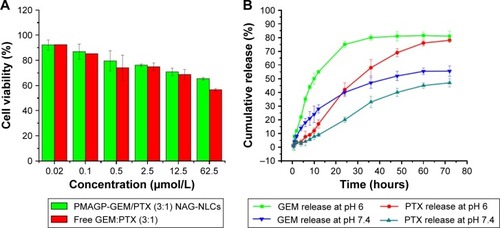
Figure 4 Confocal laser scanning microscopy images of A549 cells.
Notes: After incubation with FITC-labeled PMAGP-GEM/PTX (3:1) NLCs (A) and FITC-labeled PMAGP-GEM/PTX (3:1) NAG-NLCs (B).
Abbreviations: FITC, fluorescein isothiocyanate; PMAGP, poly(6-O-methacryloyl-d-galactopyranose); GEM, gemcitabine; PTX, paclitaxel; NLCs, nanostructured lipid carriers; NAG, N-acetyl-d-glucosamine.
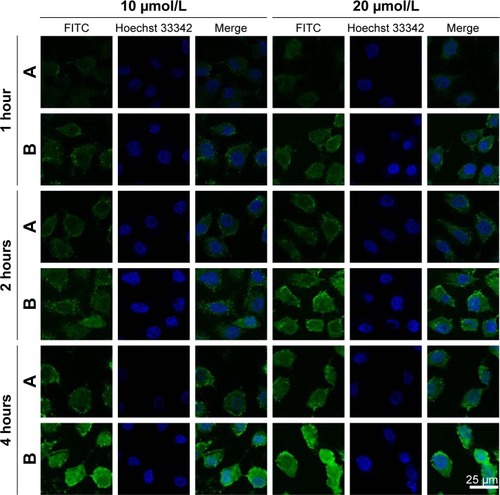
Figure 5 Cell uptake of PMAGP-GEM/PTX (3:1) NAG-NLCs and PMAGP-GEM/PTX (3:1) NAG in A549 cells analyzed by flow cytometry.
Notes: A549 cells were exposed to NAG-NLCs and NLCs with different concentrations for different time intervals. Cells incubated with only media as control. Mean fluorescence intensities show FITC intensity inside A549 cells incubated with 10 (C) and 20 (D) µmol/L, and flow-cytometry analysis (A and B) corresponds to the mean fluorescence-intensity results. Data presented as mean ± SD (n=3). **P<0.01; ***P<0.001.
Abbreviations: PMAGP, poly(6-O-methacryloyl-d-galactopyranose); GEM, gemcitabine; PTX, paclitaxel; NAG, N-acetyl-d-glucosamine; NLCs, nanostructured lipid carriers; FITC, fluorescein isothiocyanate; SD, standard deviation.
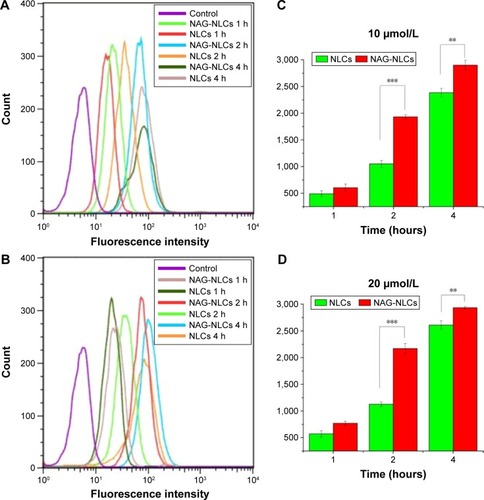
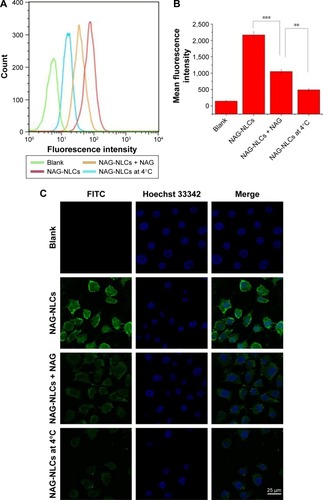
Figure 6 The competition experiments of glucose-receptor.
Notes: Blank cells, cells with FITC-labeled PMAGP-GEM/PTX (3:1) NAG-NLCs (NAG-NLCs), cells preincubated with NAG before the addition of FITC-labeled PMAGP-GEM/PTX (3:1) NAG-NLCs (NAG-NLCs + NAG), and cells with FITC-labeled PMAGP-GEM/PTX (3:1) NAG-NLCs kept at 4°C (NAG-NLCs at 4°C) in A549 cells were analyzed by flow cytometry (A, B) and confocal laser scanning microscopy (C). n=3. **P<0.01; ***P<0.001.
Abbreviations: FITC, fluorescein isothiocyanate; PMAGP, poly(6-O-methacryloyl-d-galactopyranose); GEM, gemcitabine; PTX, paclitaxel; NAG, N-acetyl-d-glucosamine; NLCs, nanostructured lipid carriers.
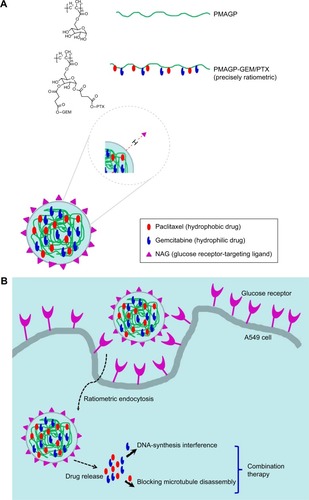
Scheme 1 Schematic illustration and intracellular performance of dual-drug load NAG-NLCs.
Notes: (A) Dual drug-loaded NAG-NLCs, of which the core consists of two drugs with ratiometric control over drug loading. (B) Intracellular performance of dual drugloaded NAG-NLCs.
Abbreviations: NAG, N-acetyl-d-glucosamine; NLCs, nanostructured lipid carriers; PMAGP, poly(6-O-methacryloyl-d-galactopyranose); GEM, gemcitabine; PTX, paclitaxel.
Scheme 2 Synthesis routes of PMAGP-GEM/PTX conjugates.
Notes: (A) PTX is conjugated with succinic anhydride through 2′-OH. (B) GEM is conjugated with succinic anhydride through hydroxyl group rather than amino group. (C) The copolymer PMaIPG was synthesized via RAFT polymerization technique and deprotected with methanoic acid. (D) 2′-succinyl-PTX must be fully reflected before 2′-succinyl-GEM added.
Abbreviations: PMAGP, poly(6-O-methacryloyl-d-galactopyranose); MaIpG, 6-O-methacryloyl-1,2;3,4-di-O-isopropylidene-d-galactopyranose; PMaIPG, poly(6-O-methacryloyl-1,2;3,4-di-O-isopropylidene-d-galactopyranose); GEM, gemcitabine; PTX, paclitaxel; RT, reaction time; TEA, triethylamine; AIBN, azobisisobutyronitrile; THF, tetrahydrofuran; DCC, dicyclohexylcarbodiimide; DMAP, dimethylaminopyridine; DMF, dimethylformamide; RAFT, reversible addition-fragmentation chain-transfer polymerization.
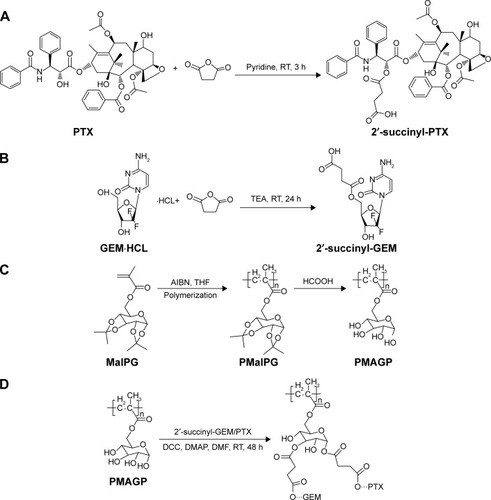
Figure S1 1H NMR spectra.
Notes: (A) PTX in DMSO-d6; (B) 2′-succinyl-PTX in CDCl3; (C) GEM·HCL in DMSO-d6; (D) 2′-succinyl-GEM in DMSO-d6; (E) PMAGP-GEM/PTX conjugates in DMSO-d6.
Abbreviations: NMR, nuclear magnetic resonance; PTX, paclitaxel; DMSO, dimethyl sulfoxide; GEM, gemcitabine; PMAGP, poly(6-O-methacryloyl-d-galactopyranose).
Figure S2 GPC traces of PMAGP (red) and PMAGP-GEM/PTX conjugates (blank).
Abbreviations: GPC, gel-permeation chromatography; PMAGP, poly(6-O-methacryloyl-d-galactopyranose); GEM, gemcitabine; PTX, paclitaxel.
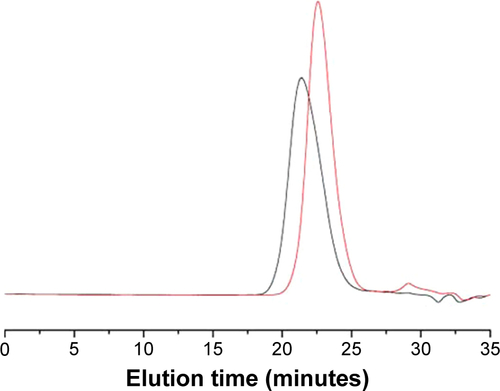
Table S1 Characterization of NLCs
