Figures & data
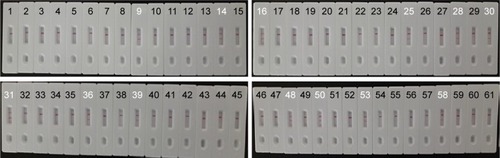
Figure 1 The outline of MCDA combined with LFB.
Notes: (A) Schematic depiction of the new cross primer (CP1*) and amplification primer (D1*). (B) Outline of MCDA with CP1* and D1*. (C) Schematic illustration of the principle of LFB for visualization of MCDA amplicons. (D) Interpretation of the results: I, positive (two red lines at the readout area) and II, negative (only the control line zone showed a red band). C1*, 5′-labeled with Dig when used in MCDA-LFB assay; CP1*, 5′-labeled with biotin when used in MCDA-LFB assay.
Abbreviations: BSA, bovine serum albumin; Dig, digoxigenin; GNPs, gold nanoparticles; LFB, lateral flow biosensor; MCDA, multiple cross displacement amplification; NC, nitrocellulose; SA, streptavidin.
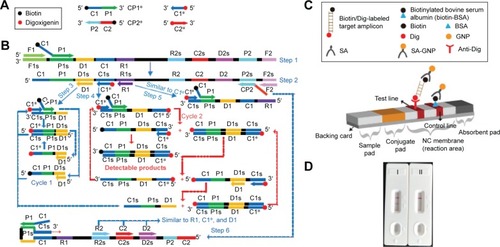
Table 1 The primers used in this study
Figure 2 Location and sequence of lmo0733 gene (Listeria monocytogenes-specific gene) used to design multiple cross displacement amplification primers.
Notes: The nucleotide sequence of the sense strand of lmo0733 is shown. Right arrows and left arrows indicate sense and complementary sequences that were used.
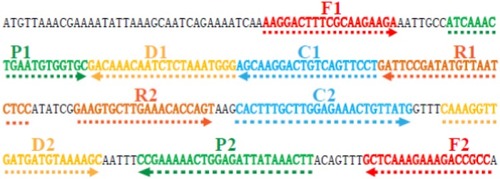
Table 2 Bacterial strains used in this study
Figure 3 Schematic representation of MCDA-LFB method compared with PCR and standard culture methods.
Abbreviations: BHI, brain heart infusion; LFB, lateral flow biosensor; MCDA, multiple cross displacement amplification; PCR, polymerase chain reaction.
Figure 4 Verification and detection of Listeria monocytogenes-MCDA products.
Notes: (A) Reaction products of L. monocytogenes-MCDA assay were visually detected by observation of the color change. (B) Agarose gel electrophoresis of L. monocytogenes-MCDA products was shown. (C) LFB applied for visual detection of L. monocytogenes-MCDA products. Tube 1/lane 1/strip 1, positive amplification of L. monocytogenes strain (EGD-e); tube 2/lane 2/strip 2, negative control of Listeria ivanovii strain (ATCCBAA-678); tube 3/lane 3/strip 3, negative control of Salmonella strain (ICDC-NPSa001); tube 4/lane 4/strip 4, blank control (DW). Lane M, DNA maker DL 100.
Abbreviations: DW, double distilled water; LFB, lateral flow biosensor; MCDA, multiple cross displacement amplification.
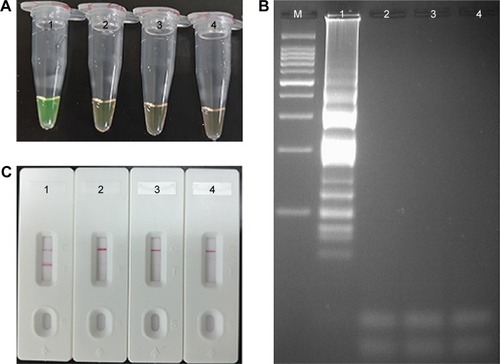
Figure 5 Optimal reaction temperature for Listeria monocytogenes-MCDA primer sets.
Notes: The standard MCDA reactions for detection of L. monocytogenes were monitored by real-time measurement of turbidity and the corresponding curves of concentrations of DNA were marked in the figures. The threshold value was 0.1 and the turbidity of >0.1 was considered to be positive. Eight kinetic graphs (A–H) were generated at various temperatures (60°C–67°C, 1°C intervals) with target pathogens DNA at the level of 10 pg per reaction. The graphs from (A) to (D) show robust amplification.
Abbreviation: MCDA, multiple cross displacement amplification.
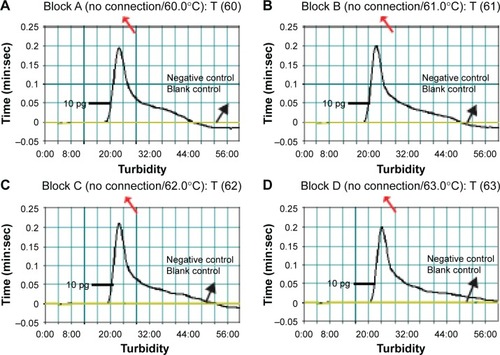
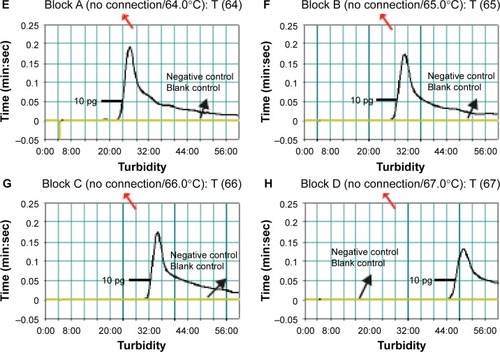
Figure 6 Analytical sensitivity of MCDA-LFB assay using serially diluted genomic DNA with Listeria monocytogenes strain EGD-e.
Notes: A total of 4 monitoring methods, including LFB (A), colorimetric indicator (FD) (B), gel electrophoresis (C), and real-time turbidity (D) were applied for analyzing the amplification products. The serial dilutions (10 ng, 10 pg, 10 fg, 1 fg, and 0.1 fg) of target templates were subjected to standard MCDA reactions. Strips (A)/tubes (B)/lanes (C)/turbidity signals (D) 1–8 represented the DNA levels of 10 ng, 10 pg, 10 fg, 1 fg, and 0.1 fg per reaction, negative control (10 pg of Listeria ivanovii genomic DNA), negative control (10 pg of Salmonella genomic DNA) and blank control (DW). The genomic DNA levels of 10 ng, 10 pg, and 10 fg per reaction produced the positive reactions.
Abbreviations: BC, blank control; DW, double distilled water; FD, fluorescent detection reagent; LFB, lateral flow biosensor; MCDA, multiple cross displacement amplification.
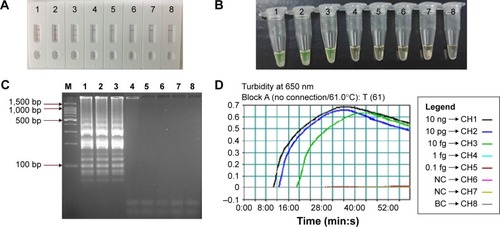
Figure 7 The optimal duration of time required for MCDA-LFB assay.
Notes: Four different reaction times (A) 10 min, (B) 15 min, (C) 20 min, and (D) 25 min were tested and compared at 61°C. Strips 1–8 represent DNA levels of 10 ng of Listeria monocytogenes templates, 10 pg of L. monocytogenes templates, 10 fg of L. monocytogenes templates, 1 fg of L. monocytogenes templates, 0.1 fg L. monocytogenes templates per tube, negative control (Listeria ivanovii, 10 pg per reaciton), negative control (Salmonella, 10 pg per reaction), and blank control (DW). The best sensitivity was seen when the amplification lasted for 20 min (C).
Abbreviations: DW, double distilled water; LFB, lateral flow biosensor; MCDA, multiple cross displacement amplification.
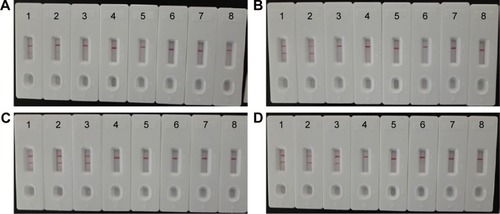
Figure 8 The specificity of MCDA-LFB assay for different strains.
Notes: The MCDA reactions were carried out using different genomic DNA templates and were monitored by means of visual format. Biosensors 1–12, Listeria monocytogenes strains of serovar 1/2a (ATCC51772), 3a (ATCC51782), 1/2c (Slcc2372), 3c (Slcc2479), 1/2b (Slcc2755), 3b (Slcc2540), 7 (NCTC10890), 4a (ATCC19114), 4c (ATCC19116), 4b (ATCC19115), 4d (ATCC19117), and 4e (ATCC19118); biosensor 13–17, Listeria ivanovii (ATCCBAA-678), Listeria innocua (ATCCBAA-680), Listeria grayi (ATCC25402), Listeria seeligeri (ATCC35897), and Listeria welshimeri (ATCC35897); biosensors 18–39, Bacillus cereus, Enteropathogenic Escherichia coli, Enterotoxigenic E. coli, Enteroaggregative E. coli, Enteroinvasive E. coli, Enterohemorrhagic E. coli, Plesiomonas shigelloides, Shigella boydii, Shigella sonneri, Shigella flexneri, Enterobacter cloacae, Enterococcus faecalis, Yersinia enterocolitica, Streptococcus pneumonia, Aeromonas hydrophil, Vibrio vulnificus, Vibrio fluvialis, Vibrio parahaemolyticus, Proteus vulgaris, Streptococcus bovis, Klebsiella pneumonia, and Salmonella enteric; and biosensor 40, blank control (DW).
Abbreviations: ATCC, American Type Culture Collection; DW, double distilled water; LFB, lateral flow biosensor; MCDA, multiple cross displacement amplification.
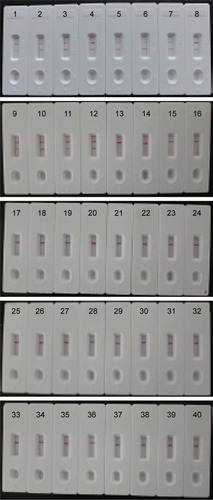
Table 3 Comparison of culture-biotechnical, MCDA-LFB, conventional PCR, for the detection of Listeria monocytogenes in raw meat samples
Figure 9 MCDA-LFB assay for the detection of Listeria monocytogenes in raw meat samples.
Notes: A total of 61 pork samples were analyzed using L. monocytogenes-MCDA-LFB assay, and 13 samples (samples 9, 14, 16, 25, 28, 30, 31, 36, 39, 48, 50, 53, and 58) were L. monocytogenes positive.
Abbreviations: LFB, lateral flow biosensor; MCDA, multiple cross displacement amplification.