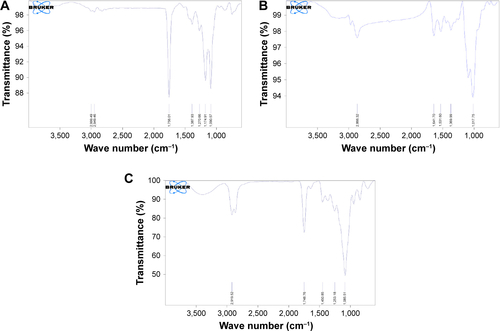
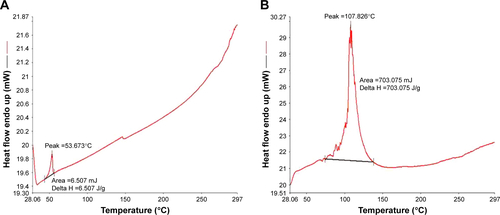
Figures & data
Table 1 Summary of experimental design, with actual and predicted values of dependent factors for NK polymeric NPs
Figure 1 3-D surface and contour plots.
Note: Interaction effects of independent variables, such as drug loading, polymer concentration, sonication time, and concentration of surfactants, on particle size (PS).
Abbreviation: PLGA, poly(lactic-co-glycolic acid) nanoparticle.
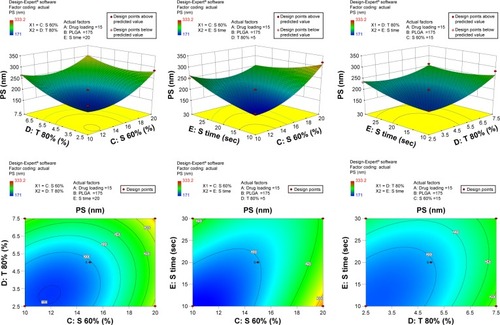
Figure 2 3-D surface and contour plots.
Note: Interaction effects of independent variables, such as drug loading, polymer concentration, sonication time, and concentration of surfactants, on polydispersity index (PDI).
Abbreviation: PLGA, poly(lactic-co-glycolic acid) nanoparticle.
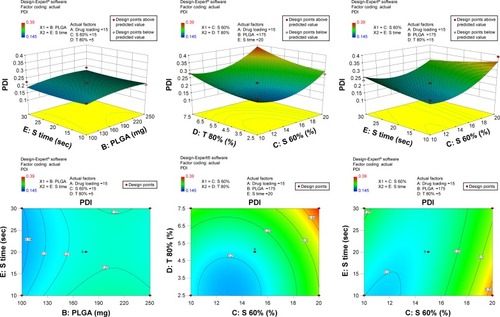
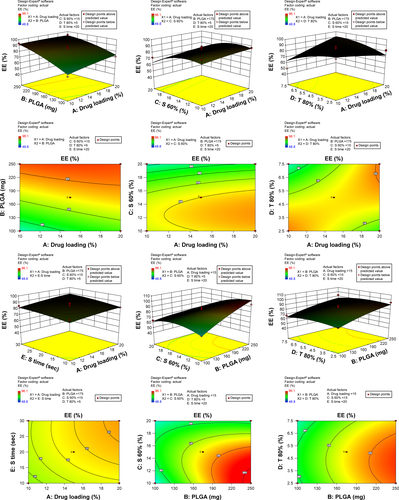
Figure 3 3-D surface and contour plots.
Note: Interaction effects of independent variables, such as drug loading, polymer concentration, sonication time and concentration of surfactants, on entrapment efficiency (EE).
Abbreviation: PLGA, poly(lactic-co-glycolic acid) nanoparticle.
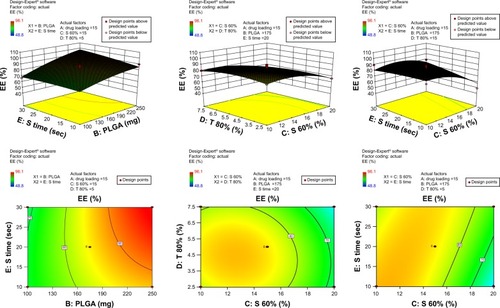
Table 2 Analysis of variance of calculated model of NK polymeric NPs
Figure 4 SEM of the optimized NPNP formulation at 10.46 K× (A) and 10.69 K× (B) magnification.
Abbreviations: SEM, scanning electron microscopy; NPNP, nattokinase–poly(lactic-co-glycolic acid) nanoparticle.
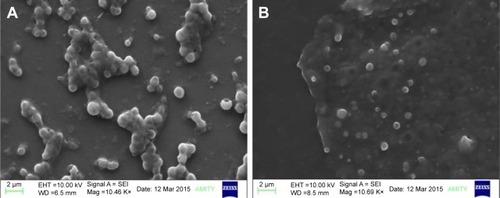
Figure 5 TEM of (A) conjugated and (B) optimized NPNP formulation.
Abbreviations: TEM, transmission electron microscopy; NPNP, nattokinase–poly(lactic-co-glycolic acid) nanoparticle; PLGA, poly(lactic-co-glycolic acid) nanoparticle.
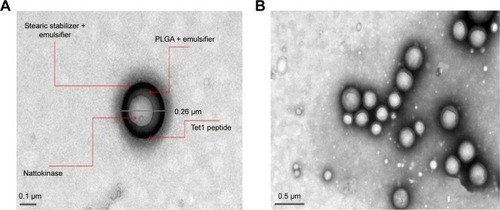
Figure 6 (A) Particle size and PDI of NKNPs; (B) particle-size distribution of NKNPs.
Abbreviations: PDI, polydispersity index; NKNPs, nattokinase–poly(lactic-co-glycolic acid) nanoparticles.
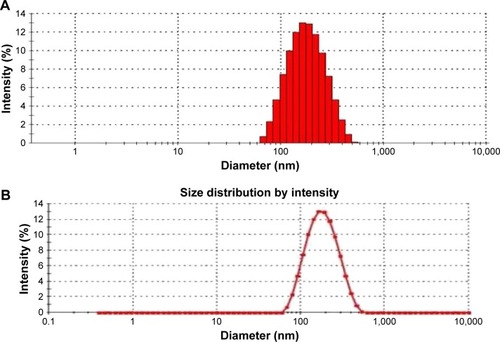
Figure 7 In vitro release of nattokinase from nattokinase polymeric nanoparticles at different time intervals (0–48 hours) using dialysis in PBS at pH 7.4.
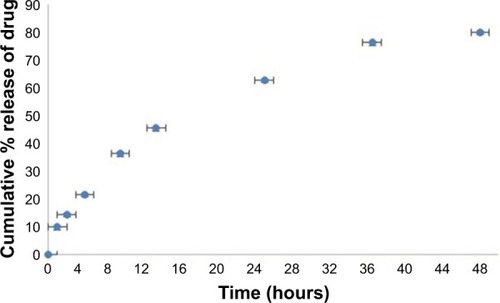
Figure 8 Fluorescence emission spectra obtained by using ThT binding assay of Aβ-40 plaque before (green line) and after digestion (24 hours) by nattokinase (red line) and NKNPs (blue line) and ThT was used as a blank (purple line) at 40°C, pH 7.
Abbreviations: ThT, thioflavin T; NK, nattokinase; PLGA, poly(lactic-co-glycolic acid); NKNPs, nattokinase–poly(lactic-co-glycolic acid) nanoparticles.
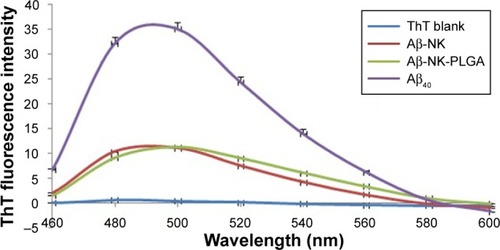
Figure 9 Scanning electron microscopy of Aβ40 plaques.
Notes: Untreated Aβ40 plaque (A and B), Aβ40 plaques digested with unformulated nattokinase for 1 h (C), Aβ40 plaques digested with unformulated nattokinase for 24 h (D), Aβ40 plaques digested with NKNPs for 1 h (E) and for 24 h (F), Aβ40 plaques digested with conjugated NKNPs for 1 h (G) and for 24 h (H).
Abbreviations: h, hours; NKNPs, nattokinase–poly(lactic-co-glycolic acid) nanoparticles.
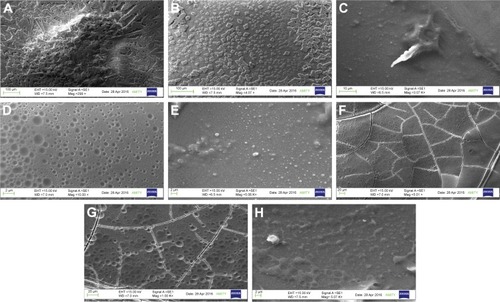
Figure 10 Invitro thrombin plaque inhibition of NK (A and D), NKNPs (B and E), and NKNPs-Tel-1 (C and F) before (A–C) and after (D–F) the preparation of NKNPs and surface modification.
Abbreviations: NK, nattokinase; NKNPs, nattokinase–poly(lactic-co-glycolic acid) nanoparticles.
Figure S1 3-D surface and contour plots.
Note: Interaction effects of independent variables, such as drug loading, polymer concentration, sonication time, and concentration of surfactants, on particle size (PS).
Abbreviation: PLGA, poly(lactic-co-glycolic acid) nanoparticle.
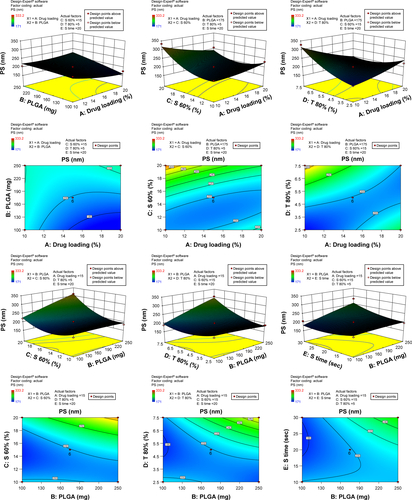
Figure S2 3-D surface and contour plots.
Note: Interaction effects of independent variables, such as drug loading, polymer concentration, sonication time, and concentration of surfactants, on polydispersity index (PDI).
Abbreviation: PLGA, poly(lactic-co-glycolic acid) nanoparticle.
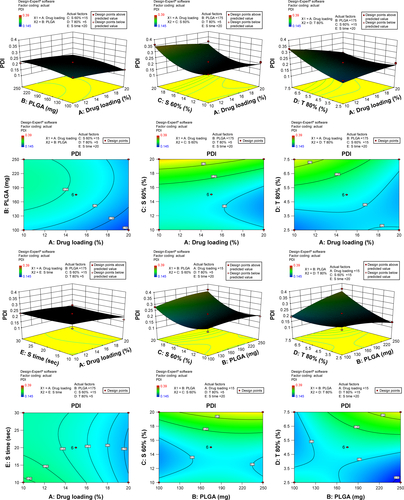
Figure S3 3-D surface and contour plots.
Note: Interaction effects of independent variables, such as drug loading, polymer concentration, sonication time, and concentration of surfactants, on entrapment efficiency (EE).
Abbreviation: PLGA, poly(lactic-co-glycolic acid) nanoparticle.