Figures & data
Figure 1 The configuration of PLGA nanofiber microchip for the high-purity isolation of CTCs.
Notes: (A) A transparent PN-NanoVelcro substrate was prepared by depositing electrospun PLGA nanofibers onto a commercial LMD glass slide (with a pre-deposited 1.2 µm thick PPS membrane). The SEM image of the electrospun PLGA nanofibers is shown. (B) A custom-designed chip holder is used to sandwich a microchip that is composed of an overlaid PDMS chaotic mixer chip. (C) Schematic representation of the chemistry of EpCAM-modified PLGA for cancer cell capture applications. (D) Single CTC isolation by laser microdissection.
Abbreviations: CTCs, circulating tumor cells; EpCAM, epithelial cell adhesion molecule; LMD, laser microdissection; PLGA, poly(lactic-co-glycolic acid); SEM, scanning electron micrograph.
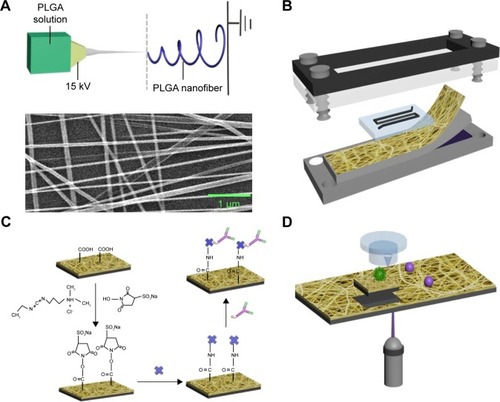
Table 1 Clinical characteristics of 72 NSCLC patients
Figure 2 Optimization and validation of the PN-NanoVelcro chip using circulating tumor cell control kit and various cell lines.
Notes: (A) The cell capture efficiency of PN-NanoVelcro chip at different electrospinning time points. (B) The cell capture efficiency of PN-NanoVelcro chip in different electrospinning flow rates. (C) The distribution of cell in different microchannels at flow rate of 1.0 mL/h. (D) Comparison of the capture performance between PN-NanoVelcro chip and four different controls: 1) PN-NanoVelcro chip with herringbone and anti-EpCAM; 2) PN-NanoVelcro chip without herringbone structure; 3) anti-EpCAM-coated PLGA thin film with electrospinning; 4) PE-NanoVelcro chip without anti-EpCAM. (E) Capture efficiencies at different spiking cell numbers ranging from 10 to 1,000/mL. (F) Capture efficiencies of different cell lines of NSCLC and WBCs.
Abbreviations: EpCAM, epithelial cell adhesion molecule; NSCLC, non-small cell lung cancer; PLGA, poly(lactic-co-glycolic acid); WBC, white blood cell.
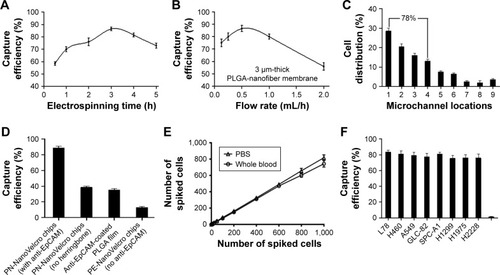
Figure 3 Identification of CTC, based on a three-color immunostaining using FITC-conjugated anti-CK, TRICT-conjugated anti-CD45 (a marker of WBC) and DAPI (nuclear specific).
Notes: NCl-H1975 cells spiked in normal blood were used as a positive control. Isolating a single CTC was performed through laser microdissection after confirming the location by immunostaining.
Abbreviations: CTC, circulating tumor cell; DAPI, 4,6-diamino-l2-phenyl indole; FITC, fluorescein isothiocyanate; TRICT, tetramethylrhodamine; WBC, white blood cell.
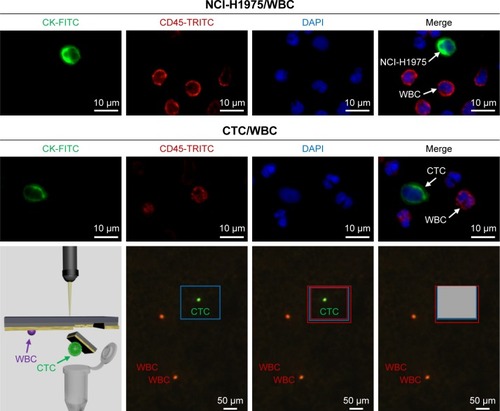
Figure 4 Concordance of EGFR mutation sites detected in primary tissues and CTCs from NSCLC patients.
Notes: (A) Landscape of somatic mutations detected in primary tissues and CTCs from 72 patients with NSCLC. Seven EGFR alterations were detected by targeted sequencing and the results were computed in the mutational landscape. (B) The Venn diagrams were used to summarize the similarity of EGFR mutations in primary tissues and CTCs from 72 NSCLC patients.
Abbreviations: CTC, circulating tumor cell; EGFR, epidermal growth factor receptor; NSCLC, non-small cell lung cancer; pri., primary tissue; WT, wild type.
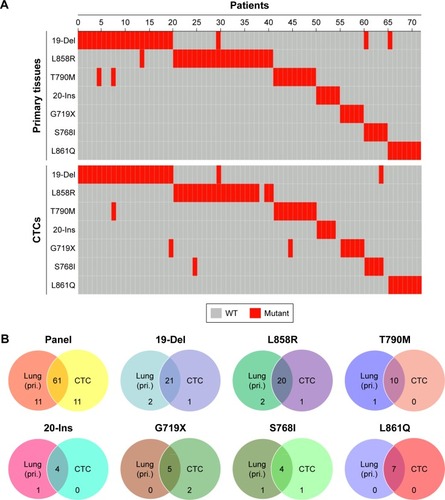
Figure 5 EGFR L858R (exon 21) and T790M (exon 20) sequencing of primary tissues and CTCs.
Notes: (A) PCR of exon 21 was successfully performed and a band of proper size was found with 2% gel electrophoresis. ARM-PCR and Sanger sequencing were used to detect the mutation sites in L858R. (B) PCR of exon 20 was successfully performed and a band of proper size was found with 2% gel electrophoresis. ARM-PCR and Sanger sequencing were used to detect the mutation sites in T790M.
Abbreviations: ARM-PCR, amplification refractory mutation system PCR; CTCs, circulating tumor cells; EGFR, epidermal growth factor receptor; PCR, polymerase chain reaction.
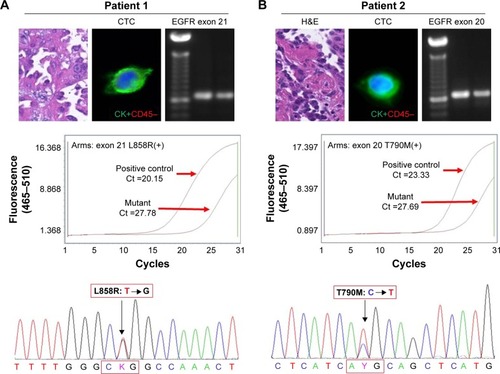
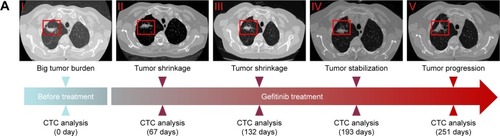
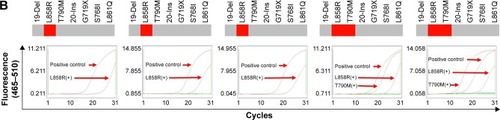
Figure 6 The relationship between acquired resistance induced by continuous gefitinib treatment and EGFR mutation.
Notes: (A) Course of the disease with CT images and treatment history and driver mutation. (B) L858R and T790M mutation detection via ARMS-PCR.
Abbreviations: ARMS-PCR, amplification refractory mutation system polymerase chain reaction; CT, computed tomography; CTC, circulating tumor cell; EGFR, epidermal growth factor receptor.