Figures & data
Table S1 Summary of recovery rate and concentrating rateTable Footnote*
Figure 1 Schematic representation of Ar/NH3 plasma-treated MNBs and their binding to anti-virus antibody.
Notes: Graphite-encapsulated MNBs were treated with ammonia plasma and modified with amino groups. SPDP was reacted with the amino group of modified MNBs (NH2-beads) at pH 7–8. Anti-rotavirus antibody or anti-dengue virus antibody was reduced using DTT, resulting in the breakage of S-S bonds and the generation of S-H groups. The S-H group of the antibody was then further reacted with SPDP-NH-MNBs. The resultant MNBs are termed antibody-integrated MNBs.
Abbreviations: DTT, dithiothreitol; MNBs, magnetic nanobeads; SPDP, N-succinimidyl 3-(2-pyridyldithio)propionate.
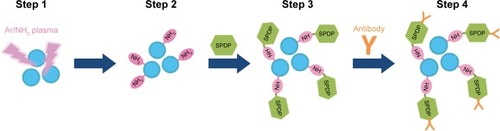
Figure 2 Schematic representation showing the capture of rotavirus using antibody-integrated MNBs.
Notes: Antibody-integrated MNBs (10 µL) were washed twice with PBS. The rotavirus-infected cell lysate (10 µL) was diluted with 500 µL of PBS and used as rotavirus suspension (Step 1). The suspension was then incubated with the MNBs for 5 minutes at room temperature (Step 2). Tubes containing the MNBs were placed in a magnetic field (Step 3). The beads were then subjected to magnetic separation, and the supernatant removed (Step 4). The beads were washed three times with PBS and resuspended in 10 µL PBS for further analysis. Steps 1–4 produced a bead fraction (BD) (10 µL antibody-integrated MNBs following incubation with PBS-diluted rotavirus-infected cell lysate), and a supernatant fraction (SP) (10 µL supernatant; following incubation and washing).
Abbreviation: MNBs, magnetic nanobeads.
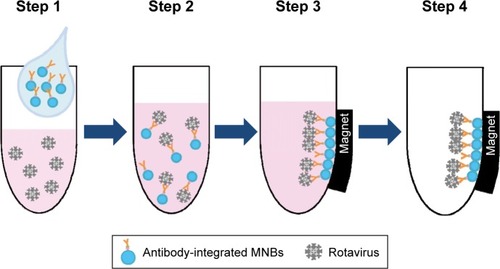
Figure 3 Detection of viral protein 6 (VP6) in rotavirus absorbed onto the antibody-integrated MNBs.
Notes: Rotavirus-infected cell lysate (10 µL) was diluted with PBS (500 µL) and incubated with antibody-integrated MNBs prior to magnetic separation. The presence of VP6 was interpreted on the basis of the presence and absence of a test line (Test). A positive control line was also included (Control). Samples were divided into the following categories: diluted rotavirus sample before incubation with the beads (BF), bead fraction after incubation with anti-rotavirus antibody-integrated MNBs (RV-BD), bead fraction after incubation with anti-dengue virus antibody-integrated MNBs (DV-BD), supernatant fraction after incubation with the anti-rotavirus antibody-integrated MNBs (RV-SP), supernatant fraction after incubation with the anti-dengue virus antibody-integrated MNBs (DV-SP), and total sample containing the same quantity of rotavirus as in 10 µL of rotavirus-infected cell lysate (total fraction, TL). All fractions were solubilized with lysis buffer and subjected to immunochromatography (ImmunoCard STAT!® Rotavirus, Meridian Bioscience Inc., Cincinnati, OH, USA) for the detection of rotavirus VP6.
Abbreviation: MNBs, magnetic nanobeads.
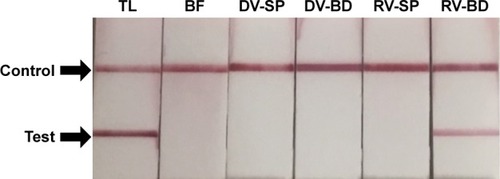
Figure 4 Detection of viral RNA of rotavirus adsorbed onto antibody-integrated MNBs.
Notes: Rotavirus-infected cell lysate (10 µL) was diluted with PBS (500 µL) and then incubated with antibody-integrated magnetic beads. After incubation, the following fractions were obtained: 1) diluted rotavirus sample before incubation with the beads (BF), 2) bead fraction after incubation with anti-rotavirus antibody-integrated MNBs (RV-BD), 3) bead fraction after incubation with anti-dengue virus antibody-integrated MNBs (DV-BD), 4) supernatant fraction after incubation with the anti-rotavirus antibody-integrated MNBs (RV-SP), 5) supernatant fraction after incubation with the anti-dengue virus antibody-integrated MNBs (DV-SP), and 6) total sample containing the same quantity of rotavirus as in 10 µL of rotavirus-infected cell lysate (total fraction, TL). Viral genomic RNA was subsequently extracted from the above fractions using a QIAamp Viral RNA mini kit and subjected to a RT-reaction. Rotavirus viral protein 7 (VP7) gene (552 bp) in the cDNA was amplified by PCR as described in Materials and methods. PCR products were analyzed by agarose gel electrophoresis (1.2% gel). The identity of the amplified products was confirmed by DNA sequencing. The left-hand lane is size marker (M), which includes DNA of 100, 200, 300, 400, 500, 600, 700, 800, 900, 1,000, 1,200, and 1,500 bp. The position of the 552 bp band for VP7 is indicated by an arrow. The NC comprised a water sample (no rotavirus) that was subjected to RT-PCR.
Abbreviations: MNBs, magnetic nanobeads; NC, negative control; RT, reverse transcription.

Figure 5 Quantitative analysis of the viral gene of rotavirus adsorbed onto the antibody-integrated MNBs.
Notes: Rotavirus-infected cell lysate (10 µL) was diluted with PBS (500 µL) and then incubated with antibody-integrated magnetic beads. After incubation, the following fractions were obtained: 1) diluted rotavirus sample before incubation with the beads (BF), 2) bead fraction after incubation with anti-rotavirus antibody-integrated MNBs (RV-BD), 3) bead fraction after incubation with anti-dengue virus antibody-integrated MNBs (DV-BD), 4) supernatant fraction after incubation with the anti-rotavirus antibody-integrated MNBs (RV-SP), 5) supernatant fraction after incubation with the anti-dengue virus antibody-integrated MNBs (DV-SP), and 6) total sample containing the same quantity of rotavirus as in 10 µL of rotavirus-infected cell lysate (total fraction, TL). Viral genomic RNA was subsequently extracted from the above fractions using a QIAamp Viral RNA mini kit and subjected to RT-reaction. The resultant cDNA was analyzed by real-time PCR using primers for the rotavirus VP7 gene as described in Materials and methods. The value of the TL sample was taken as 100%.
Abbreviations: MNBs, magnetic nanobeads; RT, reverse transcription.
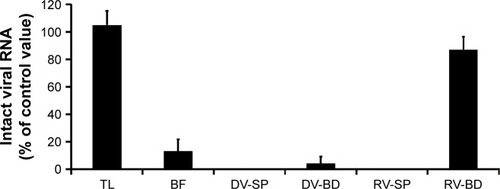
Figure 6 Infectivity in recovered rotavirus adsorbed onto the antibody-integrated MNBs.
Notes: Rotavirus-infected cell lysate (10 µL) was diluted with PBS (500 µL) and then incubated with antibody-integrated MNBs. Fractions are as follows: 1) diluted rotavirus sample before incubation with the beads (BF), 2) bead fraction after incubation with anti-rotavirus antibody-integrated MNBs (RV-BD), 3) bead fraction after incubation with anti-dengue virus antibody-integrated MNBs (DV-BD), 4) supernatant fraction after incubation with the anti-rotavirus antibody-integrated MNBs (RV-SP), 5) supernatant fraction after incubation with the anti-dengue virus antibody-integrated MNBs (DV-SP), and 6) total sample containing the same quantity of rotavirus as in 10 µL of rotavirus-infected cell lysate (total fraction, TL). All fractions were subjected to an infection analysis using MA104 cells and IFA, as described in Figure S1. The number of rotavirus-infected cells was calculated as described in Materials and methods.
Abbreviations: IFA, indirect fluorescent assay; MNBs, magnetic nanobeads.
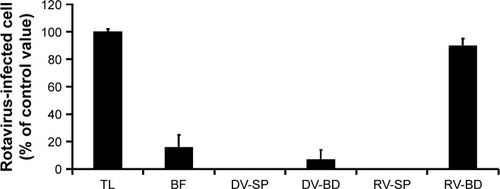
Figure S1 Recovery of infectious rotavirus adsorbed onto the antibody-integrated MNBs.
Notes: Rotavirus-infected cell lysate (10 µL) was diluted with PBS (500 µL) and then incubated with antibody-integrated MNBs. Fractions are as follows: 1) diluted rotavirus sample before incubation with the beads (BF), 2) bead fraction after incubation with anti-rotavirus antibody-integrated MNBs (RV-BD), 3) bead fraction after incubation with anti-dengue virus antibody-integrated MNBs (DV-BD), 4) supernatant fraction after incubation with the anti-rotavirus antibody-integrated MNBs (RV-SP), 5) supernatant fraction after incubation with the anti-dengue virus antibody-integrated MNBs (DV-SP), and 6) total sample containing the same quantity of rotavirus as in 10 µL of rotavirus-infected cell lysate (total fraction, TL). The fractions were added to MA104 cells, which were then cultured for 1 day. The cells were fixed with glutaraldehyde and analyzed by an IFA using anti-rotavirus antibody and DAPI solution, as described in Materials and methods. Representative fluorescence images for rotavirus (Green) (exposure time 5 seconds) and DAPI staining (Blue) (exposure time: 0.5 seconds) are shown. Scale bar=50 µm.
Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; IFA, indirect fluorescent assay; MNBs, magnetic nanobeads; RV, rotavirus.
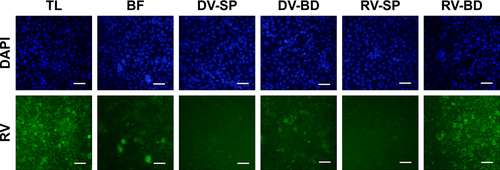
Figure S2 Comparison of rotavirus adsorption on anti-rotavirus antibody-integrated magnetic beads and magnetic beads not loaded with antibody.
Notes: Rotavirus-infected cell lysate (10 µL) was diluted with PBS (500 µL) and then incubated for 5 minutes at room temperature with either anti-rotavirus antibody-integrated magnetic beads or “plain” beads not loaded with antibody (ie, amine groups on their surface but lacking antibody). After incubation, the following fractions were obtained: 1) diluted rotavirus sample before incubation with the beads (BF), 2) supernatant fraction after incubation with “plain” beads (PL-SP), 3) bead fraction after incubation with “plain” beads (PL-BD), 4) supernatant fraction after incubation with the anti-rotavirus antibody-integrated MNBs (RV-SP), 5) bead fraction after incubation with anti-rotavirus antibody-integrated MNBs (RV-BD), and 6) total sample containing the same quantity of rotavirus as in 10 µL of rotavirus-infected cell lysate (total fraction, TL). Viral genomic RNA was subsequently extracted from each fraction using a QIAamp Viral RNA mini kit and subjected to a RT-reaction. The diluted cDNA was amplified in a reaction mixture containing Ex Taq (Takara Bio Inc.) and Ex Taq buffer as well as primers recognizing rotavirus VP7 performing 25 cycles of temperature cycling at 94°C for 1 minute, 60°C for 1 minute, and 72°C for 1 minute. Rotavirus VP7 gene (552 bp) in the cDNA was amplified by PCR as described in Materials and methods. PCR products were analyzed by agarose gel electrophoresis (1.2% gel). The identity of the amplified products was subsequently confirmed by DNA sequencing. The left-hand lane is size marker (M), which includes DNA of 100, 200, 300, 400, 500, 600, 700, 800, 900, 1,000, 1,200, and 1,500 bp. The position of the 552 bp band for VP7 is indicated by an arrow. As a negative control (NC), a water sample (ie, no rotavirus) was also subjected to RT-PCR.
Abbreviations: MNBs, magnetic nanobeads; RT, reverse transcription.
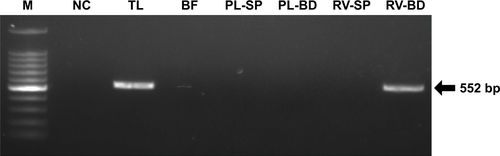
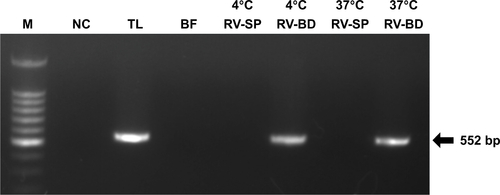
Figure S3 Comparison of rotavirus adsorption on anti-rotavirus antibody-integrated magnetic beads performed at 4°C and 37°C.
Notes: Rotavirus-infected cell lysate (10 µL) was diluted with PBS (500 µL) and then incubated for 5 minutes with anti-rotavirus antibody-integrated magnetic beads at either 4°C or 37°C. After incubation, the following fractions were obtained: 1) diluted rotavirus sample before incubation with the beads (BF), 2) supernatant fraction after incubation with the anti-rotavirus antibody-integrated MNBs at 4°C (4°C RV-SP), 3) bead fraction after incubation with anti-rotavirus antibody-integrated MNBs at 4°C (4°C RV-BD), 4) supernatant fraction after incubation with the anti-rotavirus antibody-integrated MNBs at 37°C (37°C RV-SP), 5) bead fraction after incubation with anti-rotavirus antibody-integrated MNBs at 37°C (37°C RV-BD), and 6) total sample containing the same quantity of rotavirus as in 10 µL of rotavirus-infected cell lysate (total fraction, TL). Viral genomic RNA was subsequently extracted from the above fractions using a QIAamp Viral RNA mini kit and subjected to a RT-reaction. The diluted cDNA was amplified in a reaction mixture containing Ex Taq (Takara Bio Inc.) and Ex Taq buffer as well as primers recognizing rotavirus VP7 by 30 cycles of temperature cycling at 94°C for 1 minute, 60°C for 1 minute, and 72°C for 1 minute. Rotavirus VP7 gene (552 bp) in the cDNA was amplified by PCR, as described in Materials and methods. PCR products were analyzed by agarose gel electrophoresis (1.2% gel). The identity of the amplified products was subsequently confirmed by DNA sequencing. The left-hand lane is size marker (M), which includes DNA of 100, 200, 300, 400, 500, 600, 700, 800, 900, 1,000, 1,200, and 1,500 bp. The position of the 552 bp band for VP7 is indicated by an arrow. As a NC, a water sample (ie, no rotavirus) was also subjected to RT-PCR.
Abbreviations: MNBs, magnetic nanobeads; NC, negative control; RT, reverse transcription.