Figures & data
Figure 1 Surface display of extracellular domain (ED) on the outer surface of decoy exosome. (A) Schematic illustration of vectors expressing a decoy chimera containing the extracellular domain of human tumor necrosis factor receptor 1 (hTNFR1-ED, top) or a control RFP (bottom). (B) The strategy of presenting the decoy receptor on the outer surface of exosomes using a CD63 scaffold. CD63 (blue) anchors the membrane in an M-shaped topology with its two termini located within the lumen (native exosome, Left). The control exosomes have a C-terminal GFP with or without an RFP in the second loop of CD63 (RFP-control exosome, middle). The hTNFR1-ED (purple) is fused in the second loop on the outer surface of the exosome, while a reporter GFP (green) is attached to its C-terminus inside its lumen (decoy exosome, right).
Abbreviations: CMV, cytomegalovirus promoter; N-CD63, N-terminus of CD63; hTNFR1-EC, the extracellular domain of the human TNFR1; C-CD63, C-terminus of CD63; EF-1α, elongation factor-1 promoter; Puro, puromycin resistance gene; PolyA, polyadenylation signaling sequences.
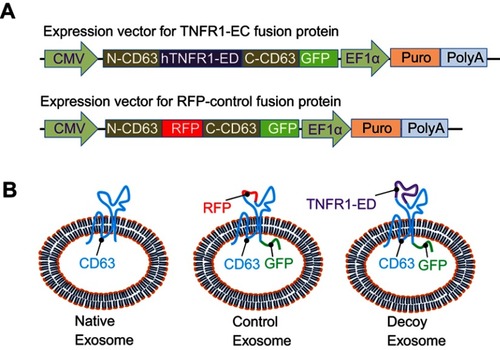
Figure 2 Establishment and characterization of stable source cell lines. (A) Scheme and timeline for the establishment of stable cell lines for production of either the CD63-RFP-GFP control exosomes or CD63-hTNFR1-GFP decoy exosomes. (B) FACS analysis of parental HEK293, CD63-RFP-GFP control, and CD63-hTNFR1-GFP stable cells. (C, D) Confocal images of the stable cell lines established by genetically transforming the parental HEK293 cells. Both GFP and RFP fusion proteins showed punctuated morphology in the cytoplasm for control CD63-RFP-GFP, while only GFP fusion proteins were expressed in CD63-hTNFR1-GFP cells. (E) Subcellular localization of the fusion proteins. Green fusion proteins were co-localized with red organelle markers for the early (1~4) or late endosomes (5~8), lysosomes (9~12), or exosomes (13~16) in the decoy exosome producing cells. Arrows indicate the organelle structures of endosome/lysosome/MVB. Scale bar: 10 µm.
Abbreviation: TLD, transmitted light detector for light imaging.
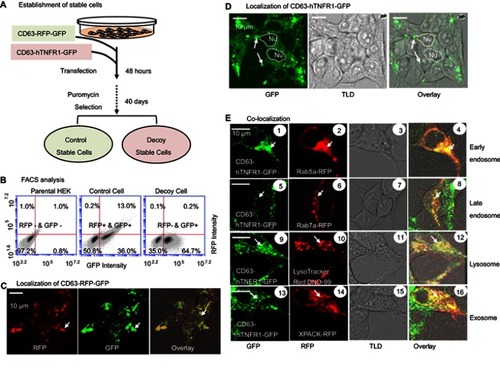
Figure 3 Exosome preparation and characterization. (A) A centrifuge-ultrafiltration-precipitation protocol used for exosome preparation. (B) Nanoparticle tracking analysis reveals exosomes in solution (left panel) along with particle size and distribution (right panel). (C) A dot-blot immuno-analysis was carried out using premade dot-blots to detect either exosomal makers, including CD63, CD81, ALIX, FLOT1, ICAM1, EpCam, ANXA5, and TSG10, or cytosolic protein GM130 for evaluating cellular contamination if nay. (D) Confocal images of isolated control and decoy exosomes.
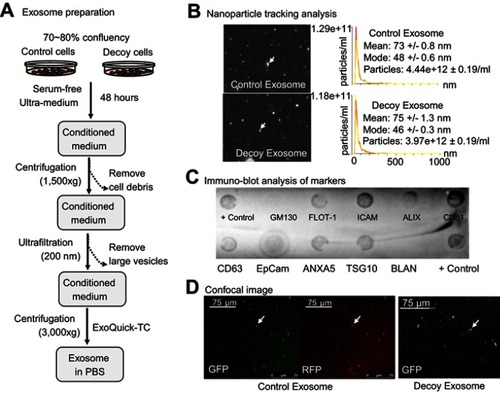
Figure 4 Efficacy of decoy exosomes on antagonizing TNFα activities in two cell models. (A) Co-culture experimental design for testing newly secreted exosomes and their antagonistic activities against TNFα. (B) Bar graph showing the specific antagonistic activities. Data are presented as mean ± SE, n=9 from 3 independent experiments. (C) Exosome treatment for testing the direct effect of decoy exosomes and their antagonistic activities against TNFα. (D) Bar graph showing the specific antagonistic activities against TNFα. Data are mean ± SE (n=6) from 2 independent experiments with two independent stable lines. The RLU was normalized to the reporter cells, which were the parental cells HEK293 cells without exosomes/engineered HEK293 cells. Column 2: Non-specific effect. Column 4: Background level activities. *Denotes P<0.05, while ***denotes P<0.001, using Student’s t-test.
Abbreviation: RLU, relative light units of luciferase activities.
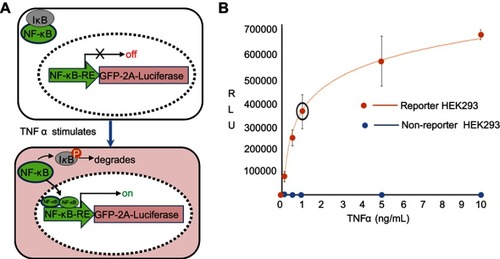
Figure S1 Reporter cell line and its dose-response to TNFα. (A) Molecular mechanism of the reporter line. A genetically encoded reporter circuit (NF-κB-RE-driven GFP and fire luciferase gene with a self-splicing 2A peptide) was incorporated into the genome of HEK293 cells. In the absence of TNFα, the transcription factor NF-κB remains in cytosol and associates with its inhibitor protein IκB. Therefore, the reporter genes has little expression due to lack of NF-κB binding to its response element (RE). In the presence of TNFα, TNFα binding to its receptor results in the phosphorylation of IκB, which leads to its degradation and a release of NF-κB. The freed NF-κB then enters into nucleus and binds the RE in the promoter of reporter, resulting in an increase of luciferase activities. (B) A dose-response of TNF α. HEK293 cells were stimulated with incremental amount of TNFα for 24 hours. Reporter cells were then collected for the luciferase activities. A dose-dependent response to TNFα was evident (red line) as compared the non-reporter parental HEK293 (blue dots).
Data availability
All data generated or analyzed in this study are included in this published article and its Supplementary information files.
