Figures & data
Figure 1 The representative image of ultrasound detected the tumor growth in liver.
Notes: Arrow indicates the tumor; length, 2.2 mm; width, 1.5 mm.
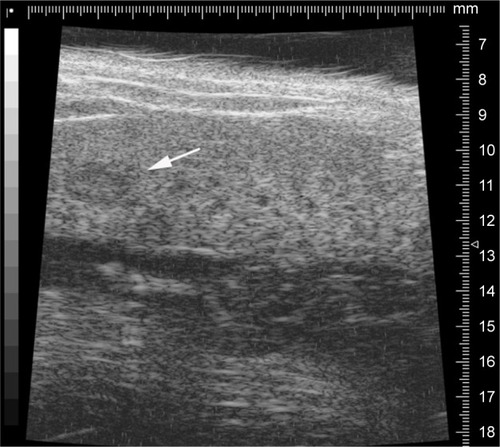
Table 1 Characterization of Adr/GNRs@PMs and Adr/GNRs@ PMs-antiEpCAM
Figure 2 Identification of Adr/GNRs@PM-antiEpCAM in Hepa1-6 cell spheroids.
Notes: Blue: DAPI positive staining of nuclei in the spheroid. Red: positive staining of antiEpCAM which were labeled Adr/GNRs@PMs.
Abbreviations: Adr, Adriamycin; EpCAM, epithelial cell adhesion molecule; GNR, gold nanorod; PM, polymeric nanomicelles.
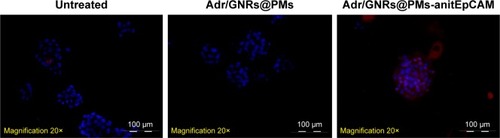
Figure 3 Cell viability by MTT assay.
Notes: (A, B) Adr/GNRs@PMs-treated Hepa1-6 cells and Hepa1-6 spheroids. (C, D) Adr/GNRs@PMs-antiEpCAM-treated Hepa1-6 cells and Hepa1-6 spheroids. (E) Temperature determined in the medium of Hepa1-6 cells with the treatments of three micelles and laser exposure. (F) Cell viability of GNRs@PMs at 4 different concentrations. (G) Cell viability of GNRs@PMs-antiEpCAM at 4 different concentrations. *P<0.05 vs control, #P<0.05.
Abbreviations: Adr, Adriamycin; C1, concentrations of 12.5 µg; C2, concentrations of 25 µg; C3, concentrations of 50 µg; C4, concentrations of 100 µg; Cont, untreated control; EpCAM, epithelial cell adhesion molecule; GNR, gold nanorod; L, laser exposure; N-SP, non-spheroids; PM, polymeric nanomicelles; s, second; SP, spheroids.
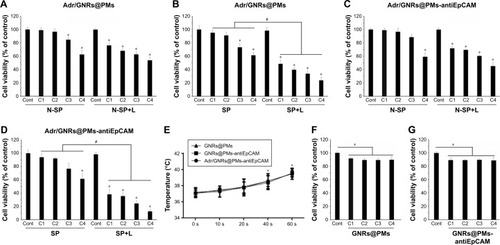
Figure 4 Identification of binary bomb-EpCAM tumor tissues of mice.
Notes: Blue: DAPI-positive staining of nuclei in the spheroid. Green: positive staining of antiEpCAM which were labeled Adr/GNRs@PMs.
Abbreviations: Adr, Adriamycin; EpCAM, epithelial cell adhesion molecule; GNR, gold nanorod; PM, polymeric nanomicelles.
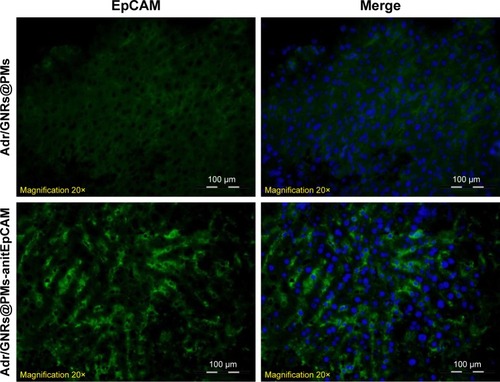
Figure 5 Histology of Adr/GNRs@PMs-antiEpCAM treatment and Adr/GNRs@PMs-antiEpCAM treatment with laser exposure.
Notes: Left: normal liver tissue and HCC cell inoculation; Right: Adr/GNRs@PMs-antiEpCAM treatment only and Adr/GNRs@PMs-antiEpCAM treatment after laser exposure. Adr/GNRs@PMs-antiEpCAM with laser exposure caused significantly cell death in the tumor tissue.
Abbreviations: Adr, Adriamycin; EpCAM, epithelial cell adhesion molecule; GNR, gold nanorod; PM, polymeric nanomicelles.
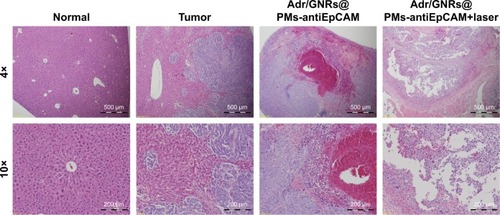
Figure 6 Upper: Quantification of HCC cell-killing in the tumor tissue by silver staining.
Notes: Middle and lower: Identification and quantification of cancer stem cell surface markers EpCAM and CD133 by immunofluorescence staining. Blue: DAPI-positive staining of nuclei in the tumor tissue. Green: positive staining of EpCAM and CD133. B (bomb): Adr/GNRs@PMs; TB (targeting bomb): Adr/GNRs@PMs-antiEpCAM. *P<0.05, **P<0.01.
Abbreviations: Adr, Adriamycin; EpCAM, epithelial cell adhesion molecule; GNR, gold nanorod; HCC, hepatocellular carcinoma; PM, polymeric nanomicelles.
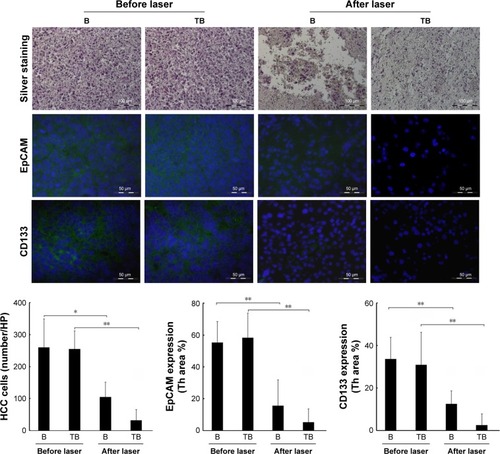
Figure 7 (A) PA spectrum of healthy liver tissue; (B and D) two different PA-US slice images acquired from healthy liver, where some ROIs (0.334 mm2) in which the PA signal was calculated are shown in (B); (C) the 3D PA-US render of healthy liver reconstructed from 176 slices of around 150 µm, the intensity of ultrasound signal in grayscale and PA records in colored bars.
Abbreviations: PA, photoacoustic; PAS, photoacoustic signal; ROI, region of interest; US, ultrasound.
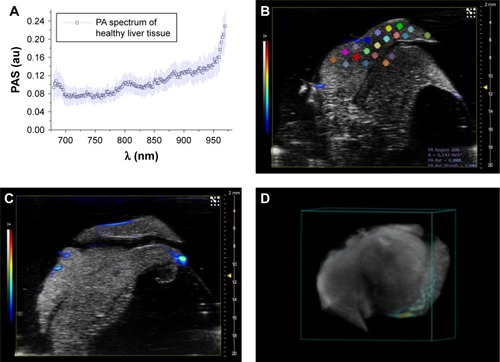
Figure 8 PA signal distribution (A, E), spectral unmixed algorithm processing (B, F), and 3D PA-US renders reconstructed by 145 slices of 150 µm thickness (C, G), and the photos of samples (D, H), for EpCAM 1 and EpCAM 2, respectively. The yellow circles were placed on the tumor regions where signal was acquired. In the PA unmixed images (B, F), the spatial distribution of nanoparticles is represented in green color, deoxy-hemoglobin chromophores in blue color, and oxy-hemoglobin chromophores in red color; gray scalebar for ultrasound signal intensity, colored scalebar for PA signal intensity.
Abbreviations: EpCAM, epithelial cell adhesion molecule; PA, photoacoustic; US, ultrasound.
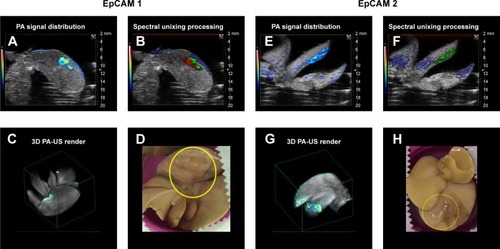
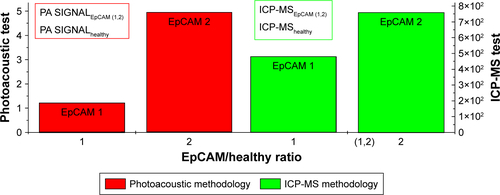
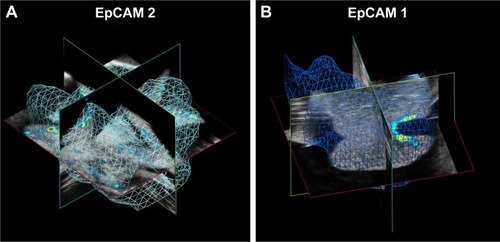
Figure 9 Volumes of calculation of averaged PA signal in (A) EpCAM 2 and (B) EpCAM 1 liver samples.
Abbreviations: EpCAM, epithelial cell adhesion molecule; PA, photoacoustic.
Scheme 1 Representative procedure for the synthesis of Adr/GNRs@PMs-antiEpCAM and TEM image of the final nanosystem.
Abbreviations: EDC, 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide; EpCAM, epithelial cell adhesion molecule; GNR, gold nanorod; NHS, N-hydroxysulfosuccinimide sodium; PM, polymeric nanomicelles; TEM, transmission electron microscopy.
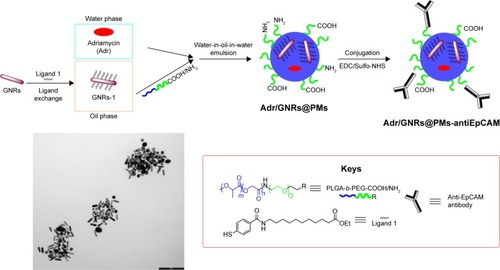
Figure S1 PA mean spectra of GNRs and that of treated liver tissue after the injection of the functionalized nanoparticles.
Abbreviations: EpCAM, epithelial cell adhesion molecule; GNR, gold nanorod; PA, photoacoustic; PAS, photoacoustic signal.
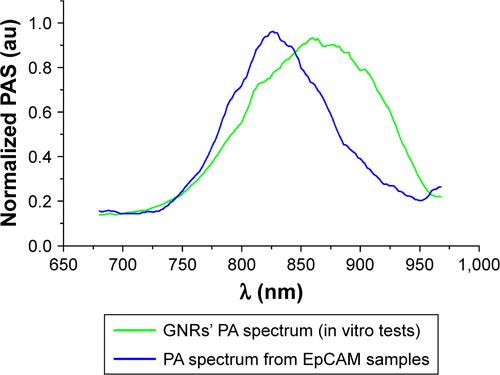
Figure S2 Spectra used for spectral unmixing processing.
Notes: The oxy- and deoxy-hemoglobin spectra had provided from default by Vevo software, and the PA spectrum of nanoparticles was calculated from in vitro tests.
Abbreviations: GNR, gold nanorod; PA, photoacoustic.
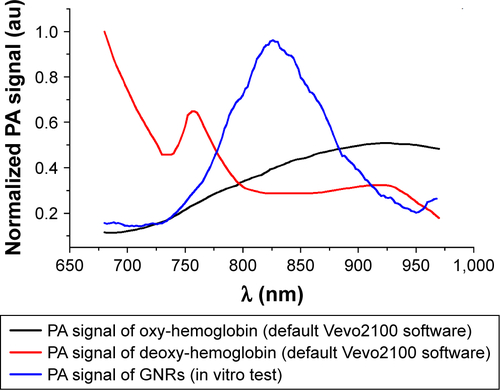
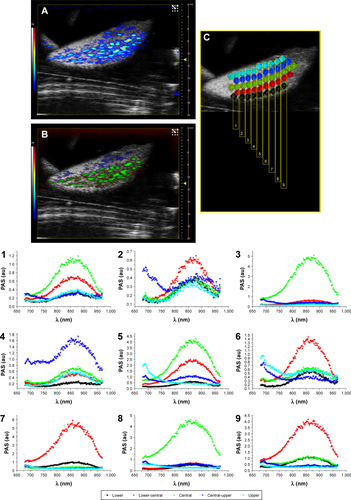
Figure S3 (A) PA-US image of a slice of liver lobe of EpCAM 2. (B) Slice of liver lobe of EpCAM 2 shown in image A, but processed with spectral unmixing. (C) The schematic of ROIs placed along 9 sections (numbered from 1 to 9) and their respective plots (also numbered from 1 to 9 and reported below) of PA signal acquired at different heights from the upper side to lower side of liver lobe.
Abbreviations: EpCAM, epithelial cell adhesion molecule; PA, photoacoustic; PAS, photoacoustic signal; ROI, region of interest; US, ultrasound.