Figures & data
Figure S1 (A) DLS measurement of the SG-CNT dispersion. (B) TEM image of SG-CNTs obtained by observation of one drop of SG-CNT dispersion on a TEM grid after drying at room temperature.
Abbreviations: DLS, dynamic light scattering; SG-CNT, super-growth carbon nanotube; TEM, transmission electron microscopy.
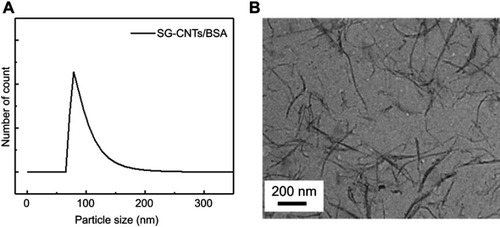
Figure 1 Degradation of SG-CNTs in RAW 264.7 macrophages. (A) DIC images of control RAW 264.7 cells incubated with SG-CNTs for 24 h (Day 0), and images taken on Day 1 and Day 7, obtained by confocal microscopy. CNTs appear as black spots. (B) The intracellular SG-CNT levels in RAW 264.7 cells at the indicated time points were estimated from the optical absorbance of the cell lysate at 750 nm. Data represent the percentages of SG-CNTs relative to the starting concentration (Day 0) and are expressed as the mean ± SD of three independent replicates. The insets show the cell lysates at Day 0 and Day 7. (C) Microscopy Raman mapping images of G-band intensities of SG-CNTs in RAW 264.7 cells fixed with glutaraldehyde at Day 0 and Day 7 (right), along with the corresponding microscopy images (left). (D) The ratio of G-band and D-band intensities for cell lysates obtained at each time point.
Abbreviations: DIC, differential interference contrast; SG-CNT, super-growth carbon nanotube.
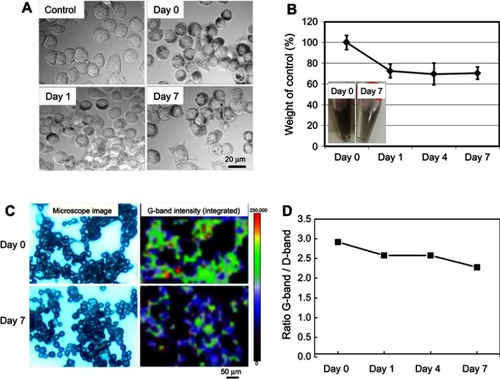
Figure 2 Degradation of SG-CNTs in Kupffer cells. (A) Confocal microscopy DIC images of control Kupffer cells and cells incubated with SG-CNTs for 24 h (Day 0), and on Day 1 and Day 7. CNTs appear as black spots. Scale bar, 20 µm. (B) The intracellular SG-CNT levels in Kupffer cells at various time points were estimated from the optical absorbance of cell lysate at 750 nm. Data represent the percentage of SG-CNTs relative to the starting concentration and are expressed as the mean ± SD of three independent replicates.
Abbreviations: DIC, differential interference contrast; SG-CNT, super-growth carbon nanotube.
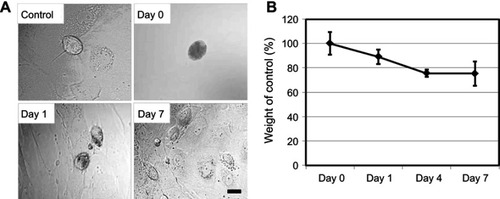
Figure S2 Degradation of ox-SG-CNTs in Kupffer cells. (A) Confocal microscopy DIC images of control Kupffer cells and that incubated with ox-SG-CNTs for 24 h (Day 0), and at Day 5 and Day 15. CNTs appear as black spots. Scale bar, 20 µm. (B) The intracellular ox-SG-CNT levels in Kupffer cells at various time points were estimated from the optical absorbance of cell lysate at 750 nm. Data represent the percentage of SG-CNTs relative to the starting concentration and are expressed as the mean ± SD of three independent replicates.
Abbreviations: DIC, differential interference contrast; ox-SG-CNT, super-growth carbon nanotubes after treatment with H2SO4/HNO3 for about 40 min at 70oC.
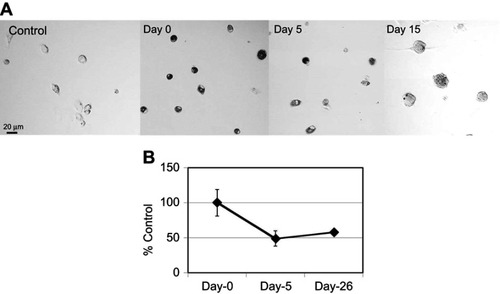
Figure 3 Cytotoxicity assay of RAW 264.7 and Kupffer cells. RAW 264.7 or Kupffer cells were incubated with SG-CNTs for 24 hrs and washed (Day 0). Cell viability was assessed after 1–7 days of further incubation without adding additional SG-CNTs to the medium. (A) WST-1 assay results. As a positive control, cells were exposed to 1 µg/mL Act D for 24 h. Data were normalized to the viability of control cells (non-CNT-treated) at each time point and represent the mean ± SD of five replicates. (B) Bradford assay results. The data were normalized to the protein concentration in control cells (non-CNT-treated) at Day 0 and represent the mean ± SD of five replicates. (C) Caspase 3/7 activity of RAW 264.7 and Kupffer cells. As a positive control, cells were treated with 1 µg/mL Act D for 3 h. Data were normalized to the activity in control cells (non-CNT-treated) at each time point and represent the mean ± SD of five replicates.
Abbreviations: Act D, actinomycin D; SG-CNT, super-growth carbon nanotube.
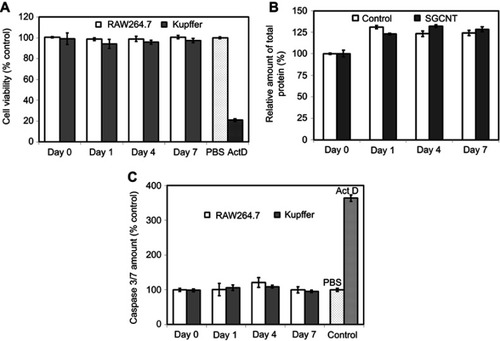
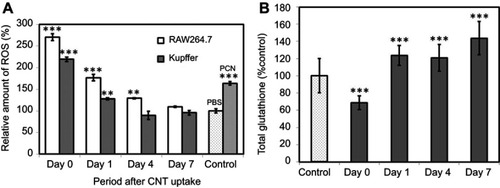
Figure 4 (A) ROS generation in RAW 264.7 and Kupffer cells. Positive control cells treated with 200 µM PCN for 3 h. Data represent the ROS level relative to that of control cells (non-CNT-treated) at each time point and are expressed as the mean ± SD of three independent replicates. (B) Total glutathione levels in RAW 264.7 macrophages after SG-CNT uptake. Data represent the glutathione level relative to that of control cells (non-CNT-treated) at each time point, and are expressed as the mean ± SD of three independent replicates. **P<0.01 and ***P<0.001 versus control cells.
Abbreviations: PCN, pyocyanin; SG-CNT, super-growth carbon nanotube.