Figures & data
Table 1 Characterization of the physico-chemical properties of the NGs
Figure 1 A reactive precursor particle approach gives access to a NG library with varying hydrophobicity and similar colloidal features.
Notes: Schematic synthesis of the NGs is depicted (A) based on precursor particles being subsequently amine functionalized (B). We used HPA that translates to PHPMA and four modified variants that each contained 80 mol-% HPA and 20 mol-% of a different hydrophobic group, ie hexylamine (HEXA-20), benzylamine (BENZA-20), linear dodecylamine (DODA-20) as well as an amine functionalized cholesteryl group (CHOLA-20).
Abbreviations: NG, nanogel; NGs, nanogels; HPA, 2-hydroxypropylamine; PHPMA, poly(N-(2-hydroxypropyl)methacrylamide); FITCA, amine functionalized fluorescein; EGDMA, ethylene glycol dimethacrylate.
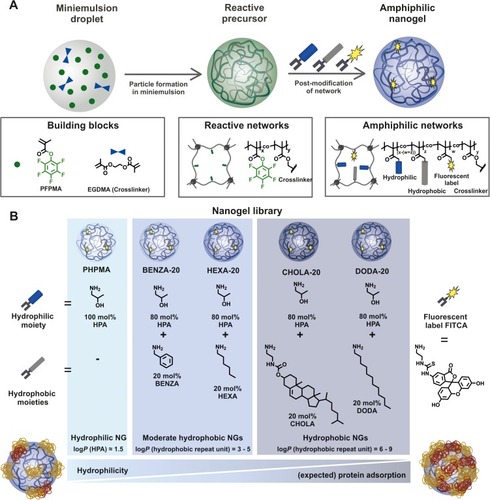
Figure 2 Contact angle measurement allows for the categorization of the NGs.
Notes: We could differentiate three groups: with PHPMA (A) as hydrophilic, BENZA-20 (B) and HEXA-20 (C) as moderate hydrophobic and CHOLA-20 (D) and DODA-20 (E) as hydrophobic.
Abbreviations: NGs, nanogels; PHPMA, poly(N-(2-hydroxypropyl)methacrylamide).
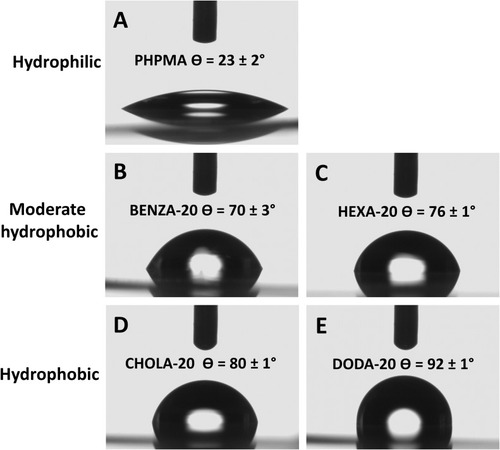
Figure 3 Illustration of size differences (Δdh) between incubation of NGs in MQ and CCM.
Notes: NGs were dispersed in MQ and CCM by vortexing, followed by 30 min sonication at RT. DLS was performed after 2 hrs.
Abbreviations: NGs, nanogels; MQ, ultrapure water; CCM, complete cell culture medium (RPMI 1640 containing 10% FBS); RT, room temperature; DLS, dynamic light scattering; PHPMA, poly(N-(2-hydroxypropyl)methacrylamide).
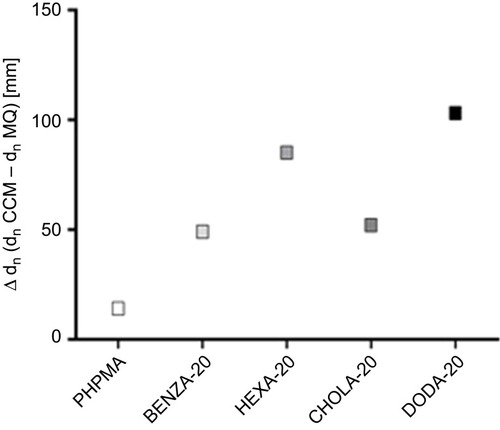
Figure 4 Viability of monocytic like THP-1 cells shows good biocompatibility of the NGs with different network hydrophobicities.
Notes: Depicted are mean values of three biological repeats and standard error of mean (SEM).
Abbreviation: NGs, nanogels.
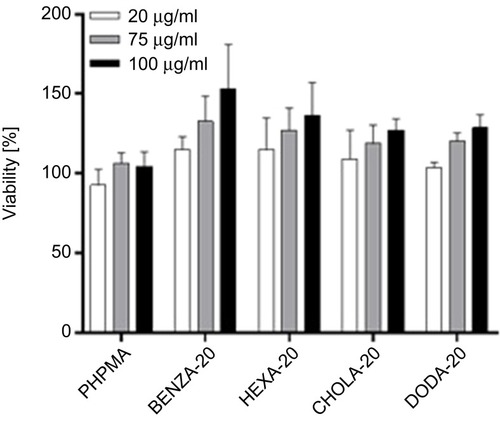
Figure 5 Differences in the protein coronas are illustrated using the spot patterns of the obtained 2DE analysis from representative NGs.
Notes: NGs (300 µg/mL) were incubated in CCM for 30 mins. Bound proteins were eluted and analyzed using 2DE analysis. Depicted are the resulting representative 2D gels for (A) PHPMA, (B) BENZA-20 and (C) DODA-20. All other 2D images are depicted in Figure S4. In comparison to blank control (no NGs), labels indicate significantly modified proteins within the respective NGs. Similarities and differences regarding the number of significant spots (ie, intensity increased >1.5, p<0.05, n=3) in each corona relative to PHPMA as a benchmark are visualized in a Venn diagram (D).
Abbreviations: 2DE, two-dimensional gel electrophoresis; NGs, nanogels; CCM, complete cell culture medium (RPMI 1640 containing 10% FBS); PHPMA, poly(N-(2-hydroxypropyl)methacrylamide).
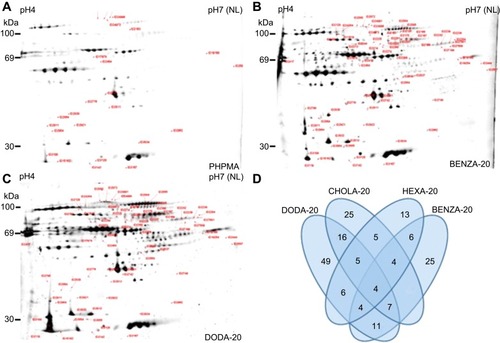
Table 2 Overview on number of corona proteins identified via LC-MS/MS compared to the hydrophilic NG PHPMA
Figure 6 Comparison of the amount of bound proteins in the corona of the respective NGs using the Top 3 approach.
Notes: Proteins in the NG coronas were identified using LC-MS/MS and abundancies were determined using the Top 3 approach. The intensity for each protein is obtained by summing up the intensities of the three most intense unique peptides for that protein. Based on that, total protein intensities for each NG were determined.
Abbreviations: NGs, nanogels; LC-MS/MS, liquid chromatography-tandem mass spectrometry; A.U., arbitrary unit; PHPMA, poly(N-(2-hydroxypropyl)methacrylamide).
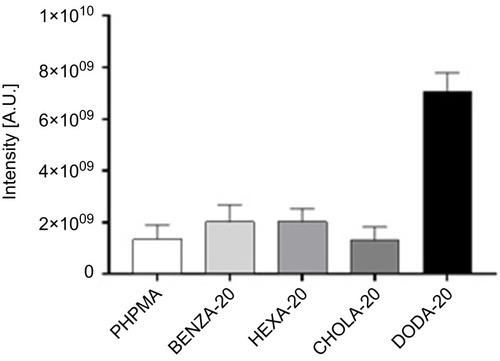
Figure 7 Impact of the various NGs upon the relative distribution of the most abundant proteins among all protein coronas.
Notes: Depicted are the 15 most abundant proteins of each single protein corona (summarized overall), which were identified via LC-MS/MS and relative percentages were calculated using the Top 3 approach. Results for each protein are given in percentage of the total bound proteins in that corona. Remaining proteins (residual % up to 100) are not depicted.
Abbreviations: NGs, nanogels; LC-MS/MS, liquid chromatography-tandem mass spectrometry; PHPMA, poly(N-(2-hydroxypropyl)methacrylamide).
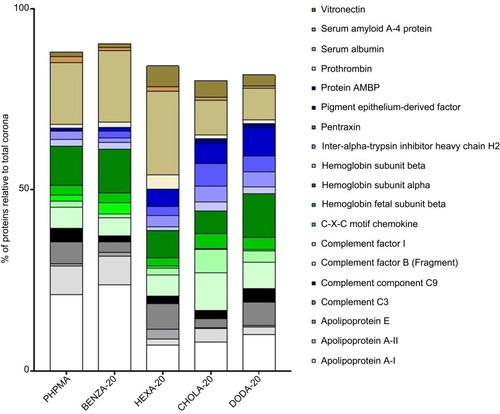
Figure 8 Cluster analysis for comparison of the NG corona proteins shows the grouping of the NGs regarding similarity within the protein corona.
Notes: Similarities and differences in the protein coronas of the NGs are visualized using a HCA. Each protein (every rectangle stands for a different identified protein) is compared to PHPMA, which serves as a benchmark particle (values are normalized) and values are transformed into a heat map. Red shows a higher intensity and green a lower intensity compared to PHPMA.
Abbreviations: NGs, nanogels; HCA, hierarchical cluster analysis; PHPMA, poly(N-(2-hydroxypropyl)methacrylamide).
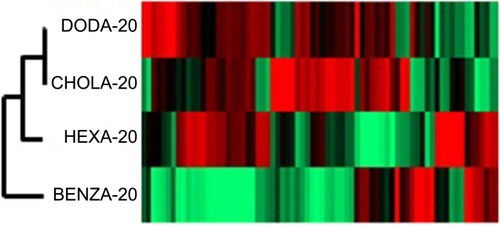
Table 3 Comparative assessment of protein coronas
Figure 9 The comparison, with regard to their intensities, of normalized expression profiles of the identified corona proteins supports the grouping of the investigated NGs.
Notes: Protein intensities were normalized (PHPMA as reference), log(2) transformed and the Z-score was calculated using Perseus Version 1.6.1.3. The expression profiles were then examined for similarities. With increasing hydrophobicity, corona proteins of the two groups of NGs, namely moderate hydrophobic (BENZA-20+HEXA-20) and hydrophobic (DODA-20+CHOLA-20), display either an increase (A) or a decrease (B) in their expression profiles. Pentraxin (C), complement factor I (D) and complement factor B (E) are depicted as examples for proteins with an increased intensity among the hydrophobic NGs compared to the moderately hydrophobic NGs. Apolipoprotein E (F) and serum albumin (G) on the other hand show a decreased intensity among the hydrophobic NGs compared to the moderate NGs.
Abbreviations: NGs, nanogels; A.U., arbitrary unit; PHPMA, poly(N-(2-hydroxypropyl)methacrylamide).
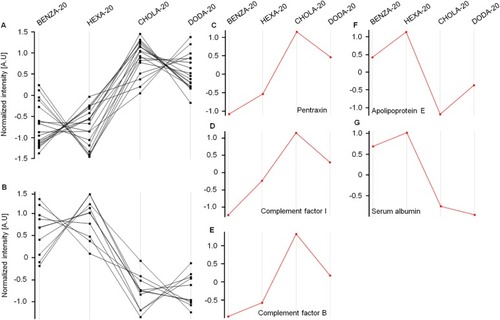
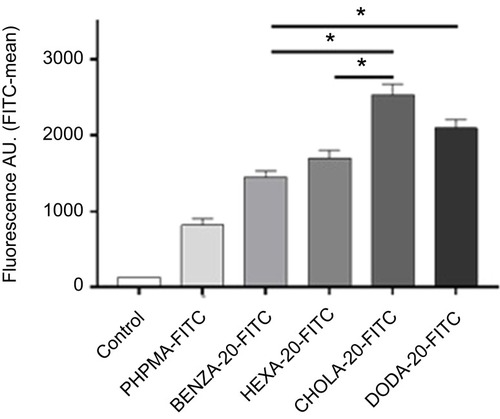
Figure 10 Flow cytometry data reveal a correlation between the uptake of NGs in human monocytic like THP-1 cells and the hydrophobicity of NGs.
Notes: NGs were pre-incubated for 2 hrs with CCM containing 10% FBS. After 2 hrs of incubation with THP-1 cells in an incubator (37°C, 5% CO2) uptake was measured using flow cytometry. Mean fluorescence values including SEM (n=3 biological repeats) as bars with significance (*P<0.0005) are depicted. All four NGs were significantly (p<0.0005) increased in uptake versus the control PHPMA (not depicted).
Abbreviations: NGs, nanogels; CCM, complete cell culture medium (RPMI 1640 with 10% FBS); SEM, standard error of mean; A.U., arbitrary unit; PHPMA, poly(N-(2-hydroxypropyl)methacrylamide).